








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 112-122.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0923
• 研究报告 • 上一篇
收稿日期:2024-09-24
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
薛飞,男,博士,教授,研究方向 :棉花生物育种;E-mail: xuefei@shzu.edu.cn作者简介:冯小康,男,硕士研究生,研究方向 :作物遗传育种;E-mail: 2364430810@qq.com
基金资助:
FENG Xiao-kang( ), LIANG Qian, WANG Xue-feng, SUN Jie, XUE Fei(
), LIANG Qian, WANG Xue-feng, SUN Jie, XUE Fei( )
)
Received:2024-09-24
Published:2025-03-26
Online:2025-03-20
摘要:
目的 SEC1复合体是位于类囊体上的蛋白质转运系统,由SCY1、SECE1和SECA1共同组成。SEC1系统参与类囊体蛋白的转运和整合,对维持叶绿体结构的稳定起着非常重要的作用。利用生物信息学从陆地棉全基因组中鉴定SEC1复合体基因,旨在为光合机制研究和棉花高产抗逆育种提供新的基因资源。 方法 基于拟南芥SEC1系统组分基因蛋白序列,从陆地棉基因组中鉴定棉花SEC1系统组分,分析该系统组分各基因的理化性质、跨膜结构、蛋白互作、启动子元件及表达模式。利用VIGS和透射电镜技术,研究叶绿体主要通道蛋白组分GhSCY1对叶绿素合成、叶绿体结构和形态的影响。 结果 在陆地棉中鉴定出了SEC1转运系统的组分,GhSCY1A/D、GhSECE1A/D和GhSECA1A/D。GhSCY1A/D含有1个SecY保守结构域,7个跨膜结构域,GhSECE1D只含有1个跨膜结构域。预测GhSCY1A/D与GhSECE1D在SEC1转运系统中共同形成叶绿体跨膜通道蛋白。GhSECA1A/D拥有1个SecA结构域,不含跨膜结构域,主要为蛋白转运过程提供能量。SEC1系统各基因在叶片中的表达量较高,且能够参与温度胁迫的响应。沉默GhSCY1基因(GhSCY1A和GhSCY1D)后,棉花叶片出现斑驳的淡黄色表型,叶绿素含量明显下降,叶片叶绿体结构和形态异常,没有淀粉粒累积。 结论 棉花SEC1系统分别由GhSCY1A/D、GhSECE1A/D和GhSECA1A/D构成,其中GhSCY1A/D基因在维持叶绿体结构和形态及叶绿素生物合成方面发挥重要作用。
冯小康, 梁倩, 王学峰, 孙杰, 薛飞. 棉花SEC1复合体组分的鉴定与GhSCY1基因功能验证[J]. 生物技术通报, 2025, 41(3): 112-122.
FENG Xiao-kang, LIANG Qian, WANG Xue-feng, SUN Jie, XUE Fei. Identification of SEC1 Complex Components and Functional Validation of the GhSCY1 Gene in Cotton[J]. Biotechnology Bulletin, 2025, 41(3): 112-122.
| 名称 Name | 序列 Sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| V-GhSCY1-F | VIGS | |
| V-GhSCY1-R | ||
| q-GhSCY1-F | GTATCAACCCAGACGAGTCTTCAG | RT-qPCR |
| q-GhSCY1-R | AACTCCAAGACGAGATAAAGCC |
表1 所用相关的引物序列
Table 1 Sequences of relevant primers used
| 名称 Name | 序列 Sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| V-GhSCY1-F | VIGS | |
| V-GhSCY1-R | ||
| q-GhSCY1-F | GTATCAACCCAGACGAGTCTTCAG | RT-qPCR |
| q-GhSCY1-R | AACTCCAAGACGAGATAAAGCC |

图1 棉花SEC1系统组分进化分析与鉴定A:GhSCY1(GH_A05G0068和GH_D05G0074)的进化分析;B:GhSECA1(GH_A12G1366和GH_D12G1384)的进化分析;C:GhSECE1(GH_A09G2370和GH_D09G2311)的进化分析;D:SEC1组分保守结构域分析。图1-A-C中蛋白质序列来源于拟南芥、玉米、水稻、烟草、雷蒙德氏棉、瑟伯氏棉、黄褐棉、海岛棉、陆地棉、亚洲棉、毛棉和达尔文棉;红色符号表示内部节点符号,表示的是假定祖先;绿色符号表示是位于树的最末端的节点(叶节点),代表基因
Fig. 1 Identification and evolutionary analysis of SEC1 system in cotton (Gossypium hirsutum)A: Evolutionary analysis of GhSCY1 (GH_A05G0068 and GH_D05G0074). B: Evolutionary analysis of GhSECA1 (GH_A12G1366 and GH_D12G1384). C: Evolutionary analysis of GhSECE1 (GH_A09G2370 and GH_D09G2311). D: Analysis of conserved domains in the SEC1 component. The protein sequences in Fig.1 A-C are derived from Arabidopsis thaliana, Zea mays, Oryza sativa, Nicotiana attenuata, Gossypium raimondii, Gossypium thurberi, Gossypium mustelinum, Gossypium barbadense, Gossypium hirsutum, Gossypium arboreum, Gossypium tomentosum, and Gossypium darwinii. The red symbols indicate internal node symbols, which denote the presumed ancestors; the green symbols indicate the nodes at the very end of the tree (leaf nodes), representing genes
基因名称 Gene name | 基因ID Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 亲水性 GRAVY | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| GhSCY1A | GH_A05G0068 | 542 | 58.612 73 | 9.48 | 37.39 | 0.258 | 叶绿体Chloroplast |
| GhSCY1D | GH_D05G0074 | 542 | 58.709 83 | 9.56 | 36.39 | 0.244 | 叶绿体Chloroplast |
| GhSECA1A | GH_A12G1366 | 1 037 | 116.944 51 | 6.23 | 42.82 | -0.368 | 叶绿体Chloroplast |
| GhSECA1D | GH_D12G1384 | 1 037 | 116.993 61 | 6.33 | 43.19 | -0.366 | 叶绿体Chloroplast |
| GhSECE1A | GH_A09G2370 | 174 | 18.592 99 | 5.53 | 46.77 | -0.208 | 叶绿体Chloroplast |
| GhSECE1D | GH_D09G2311 | 174 | 18.489 82 | 5.34 | 50.07 | -0.208 | 叶绿体Chloroplast |
表2 棉花SEC1系统理化性质及亚细胞定位分析
Table 2 Analysis of physicochemical properties and subcellular localization of the SEC1 system in cotton
基因名称 Gene name | 基因ID Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 亲水性 GRAVY | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| GhSCY1A | GH_A05G0068 | 542 | 58.612 73 | 9.48 | 37.39 | 0.258 | 叶绿体Chloroplast |
| GhSCY1D | GH_D05G0074 | 542 | 58.709 83 | 9.56 | 36.39 | 0.244 | 叶绿体Chloroplast |
| GhSECA1A | GH_A12G1366 | 1 037 | 116.944 51 | 6.23 | 42.82 | -0.368 | 叶绿体Chloroplast |
| GhSECA1D | GH_D12G1384 | 1 037 | 116.993 61 | 6.33 | 43.19 | -0.366 | 叶绿体Chloroplast |
| GhSECE1A | GH_A09G2370 | 174 | 18.592 99 | 5.53 | 46.77 | -0.208 | 叶绿体Chloroplast |
| GhSECE1D | GH_D09G2311 | 174 | 18.489 82 | 5.34 | 50.07 | -0.208 | 叶绿体Chloroplast |
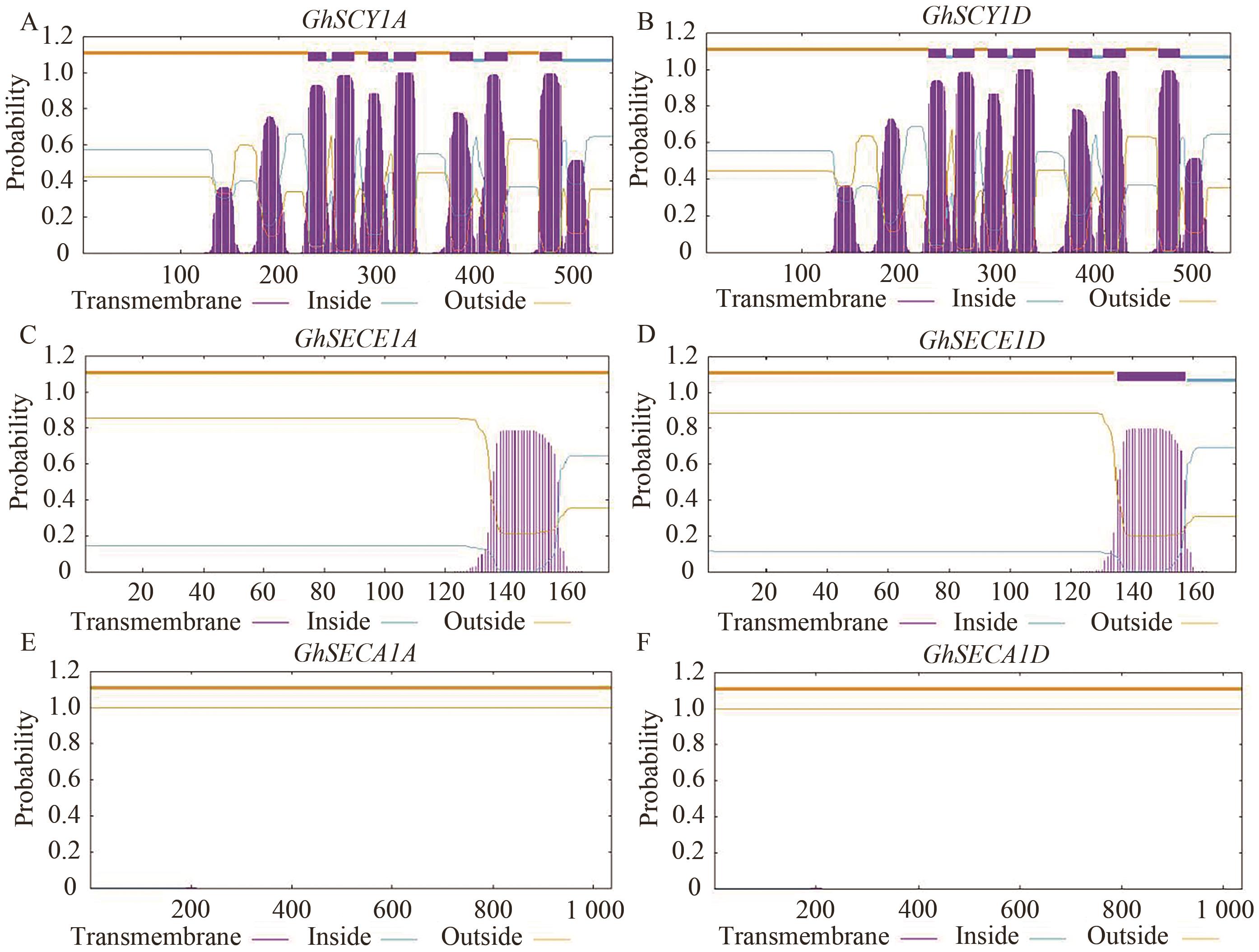
图2 棉花SEC1系统组分跨膜结构分析图中紫色方框个数表示跨膜结构的数量;A‒F分别表示GhSCY1A含有7个跨膜结构、GhSCY1D含有7个跨膜结构、GhSECE1A不含跨膜结构、GhSECE1D含有1个跨膜结构、GhSECA1A和GhSECA1D不含跨膜结构
Fig. 2 Analysis of the transmembrane structure of the SEC1 systemic component in cottonThe number of purple squares in the figure indicates the number of transmembrane structures; A‒F indicate that GhSCY1A contains 7 transmembrane structures, GhSCY1D contains 7 transmembrane structures, GhSECE1A does not contain any transmembrane structures, GhSECE1D contains 1 transmembrane structure, and both GhSECA1A and GhSECA1D do not contain transmembrane structures

图3 棉花SEC1系统组分三维结构互作分析A:GhSCY1、GhSECE1 和GhSECA1之间的互作;B:GhSCY1与GhSECA1的互作;C:GhSCY1与GhSECE1的互作;D:GhSECE1 和GhSECA1之间的互作;绿色:GhSCY1蛋白结构;蓝色:GhSECE1蛋白结构;黄色:GhSECA1蛋白结构;红色:蛋白间的互作区域
Fig. 3 Analysis on three-dimensional structural interactions analysis of SEC1 system components in cottonA: Interaction between GhSCY1, GhSECE1 and GhSECA1. B: Interaction between GhSCY1 and GhSECA1. C: Interaction between GhSCY1 and GhSECE1. D: Intercropping between GhSECE1 and GhSECA1. Green color: Protein structure of GhSCY1; blue color: protein structure of GhSECE1; yellow color: protein structure of GhSECA1; red color: region of interactions between proteins

图7 棉花GhSCY1A/D基因沉默后的表型、表达量及色素含量A:GhCHLI沉默后表型;B:TRV:00植株和TRV:GhSCY1植株沉默后的整株表型;C:GhSCY1在TRV:00和TRV:GhSCY1植株中的相对表达量;D:TRV:00和TRV:GhSCY1沉默后叶片表型;E:TRV:00和TRV:GhSCY1沉默后总叶绿素含量;F:TRV:00和TRV:GhSCY1沉默后的胡萝卜素含量;***:在0.01水平上存在显著差异,****:在0.001水平上存在显著差异
Fig. 7 Phenotype, expression and pigment content after silencing of GhSCY1A/D gene in cottonA: Phenotype of GhCHLI silencing. B: Whole plant phenotype after silencing with TRV:00 and TRV:GhSCY1. C: Relative expressions of GhSCY1 in TRV:00 and TRV:GhSCY1 plants. D: Leaf phenotype after silencing with TRV:00 and TRV:GhSCY1. E: Total chlorophyll content after silencing with TRV:00 and TRV:GhSCY1. F: Carotenoid content after silencing with TRV:00 and TRV:GhSCY1. ***: Significant difference at the 0.01 level, ****: significant difference at the 0.001 level
| 1 | Chen YZ, Fu MC, Li H, et al. High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system [J]. Plant Biotechnol J, 2021, 19(3): 424-426. |
| 2 | Liu RX, Xiao XH, Gong JW, et al. QTL mapping for plant height and fruit branch number based on RIL population of upland cotton [J]. J Cotton Res, 2020, 3(1): 5. |
| 3 | Paterson AH, Wendel JF, Gundlach H, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres [J]. Nature, 2012, 492(7429): 423-427. |
| 4 | 许子怡, 程行, 沈奇, 等. 水稻黄绿叶突变体ygl3的鉴定与基因功能分析 [J]. 中国农业科学, 2021, 54(15): 3149-3157. |
| Xu ZY, Cheng X, Shen Q, et al. Identification and gene functional analysis of yellow green leaf mutant ygl3 in rice [J]. Sci Agric Sin, 2021, 54(15): 3149-3157. | |
| 5 | Jung KH, Hur J, Ryu CH, et al. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system [J]. Plant Cell Physiol, 2003, 44(5): 463-472. |
| 6 | 孙小秋, 王兵, 肖云华, 等. 水稻ygl98黄绿叶突变基因的精细定位与遗传分析 [J]. 作物学报, 2011, 37(6): 991-997. |
| Sun XQ, Wang B, Xiao YH, et al. Genetic analysis and fine-mapping of ygl98 yellow-green leaf gene in rice [J]. Acta Agron Sin, 2011, 37(6): 991-997. | |
| 7 | Mao GZ, Ma Q, Wei HL, et al. Fine mapping and candidate gene analysis of the virescent gene v1 in Upland cotton (Gossypium hirsutum) [J]. Mol Genet Genomics, 2018, 293(1): 249-264. |
| 8 | Zhu JK, Chen JD, Gao FK, et al. Rapid mapping and cloning of the virescent-1 gene in cotton by bulked segregant analysis-next generation sequencing and virus-induced gene silencing strategies [J]. J Exp Bot, 2017, 68(15): 4125-4135. |
| 9 | Shim KC, Kang YN, Song JH, et al. A frameshift mutation in the Mg-chelatase I subunit gene OsCHLI is associated with a lethal chlorophyll-deficient, yellow seedling phenotype in rice [J]. Plants (Basel), 2023, 12(15): 2831. |
| 10 | Fang GN, Yang SL, Ruan BP, et al. Isolation of TSCD11 gene for early chloroplast development under high temperature in rice [J]. Rice (N Y), 2020, 13(1): 49. |
| 11 | Huo YZ, Cheng MX, Tang MJ, et al. GhCTSF1, a short PPR protein with a conserved role in chloroplast development and photosynthesis, participates in intron splicing of rpoC1 and ycf3-2 transcripts in cotton [J]. Plant Commun, 2024, 5(6): 100858. |
| 12 | Roose JL, Frankel LK, Bricker TM. The PsbP domain protein 1 functions in the assembly of lumenal domains in photosystem I [J]. J Biol Chem, 2014, 289(34): 23776-23785. |
| 13 | Liu J, Yang HX, Lu QT, et al. PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis [J]. Plant Cell, 2012, 24(12): 4992-5006. |
| 14 | Lu HJ, Xiao YY, Liu YX, et al. Integrative transcriptomics and proteomics analysis of a cotton mutant yl1 with a chlorophyll-reduced leaf [J]. Plants (Basel), 2024, 13(13): 1789. |
| 15 | 李劲宇, 安传敬, 刘小敏, 等. 叶绿体分裂分子机制研究进展 [J]. 植物生理学报, 2016, 52(11): 1733-1744. |
| Li JY, An CJ, Liu XM, et al. Recent progress in the molecular mechanism of chloroplast division [J]. Plant Physiol J, 2016, 52(11): 1733-1744. | |
| 16 | Xu XM, Ouyang M, Lu DD, et al. Protein sorting within chloroplasts [J]. Trends Cell Biol, 2021, 31(1): 9-16. |
| 17 | Richardson LGL, Singhal R, Schnell DJ. The integration of chloroplast protein targeting with plant developmental and stress responses [J]. BMC Biol, 2017, 15(1): 118. |
| 18 | Bauer J, Chen K, Hiltbunner A, et al. The major protein import receptor of plastids is essential for chloroplast biogenesis [J]. Nature, 2000, 403(6766): 203-207. |
| 19 | Hust B, Gutensohn M. Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana [J]. Plant Biol, 2006, 8(1): 18-30. |
| 20 | Jarvis P, Chen LJ, Li H, et al. An Arabidopsis mutant defective in the plastid general protein import apparatus [J]. Science, 1998, 282(5386): 100-103. |
| 21 | Kikuchi S, Bédard J, Hirano M, et al. Uncovering the protein translocon at the chloroplast inner envelope membrane [J]. Science, 2013, 339(6119): 571-574. |
| 22 | Nakai M, Sugita D, Omata T, et al. Sec-Y protein is localized in both the cytoplasmic and thylakoid membranes in the Cyanobacterium synechococcus PCC7942 [J]. Biochem Biophys Res Commun, 1993, 193(1): 228-234. |
| 23 | Williams-Carrier R, Stiffler N, Belcher S, et al. Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize [J]. Plant J, 2010, 63(1): 167-177. |
| 24 | Zoschke R, Bock R. Chloroplast translation: structural and functional organization, operational control, and regulation [J]. Plant Cell, 2018, 30(4): 745-770. |
| 25 | Liu D, Wu ZM, Hou L. Loss-of-function mutation in SCY1 triggers chloroplast-to-nucleus retrograde signaling in Arabidopsis thaliana [J]. Biol Plant, 2015, 59(3): 469-476. |
| 26 | Yuan J, Henry R, McCaffery M, et al. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting [J]. Science, 1994, 266(5186): 796-798. |
| 27 | Nakai M, Goto A, Nohara T, et al. Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein transport [J]. J Biol Chem, 1994, 269(50): 31338-31341. |
| 28 | Schuenemann D, Amin P, Hartmann E, et al. Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex [J]. J Biol Chem, 1999, 274(17): 12177-12182. |
| 29 | Ballabani G, Forough M, Kessler F, et al. The journey of preproteins across the chloroplast membrane systems [J]. Front Physiol, 2023, 14: 1213866. |
| 30 | Fernandez DE. Two paths diverged in the stroma: targeting to dual SEC translocase systems in chloroplasts [J]. Photosynth Res, 2018, 138(3): 277-287. |
| 31 | Skalitzky CA, Martin JR, Harwood JH, et al. Plastids contain a second sec translocase system with essential functions[J]. Plant Physiol, 2011, 155(1): 354-369. |
| 32 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 33 | Lichtenthaler HK. [34]chlorophylls and carotenoids: pigments of photosynthetic biomembranes [M]//Methods in Enzymology. Amsterdam: Elsevier, 1987: 350-382. |
| 34 | 程谟桢. 番茄黄化突变体ym的黄化迟熟机制与关键基因Slym1鉴定 [D]. 哈尔滨: 东北农业大学, 2021. |
| Cheng MZ. Mechanism of yellowing late ripening of tomato mutant ym and identification of key gene Slym1 [D]. Harbin: Northeast Agricultural University, 2021. | |
| 35 | Gao LL, Hong ZH, Wang YS, et al. Chloroplast proteostasis: a story of birth, life, and death [J]. Plant Commun, 2023, 4(1): 100424. |
| 36 | Zhang H, Zhou JF, Kan Y, et al. A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance [J]. Science, 2022, 376(6599): 1293-1300. |
| 37 | 庄焜扬. 温度胁迫下番茄叶绿体与细胞核双定位WHIRLY1蛋白的功能分析 [D]. 泰安: 山东农业大学, 2020. |
| Zhuang KY. Functional analysis of WHIRLY1 protein with double localization in chloroplast and nucleus of tomato under temperature stress [D]. Tai'an: Shandong Agricultural University, 2020. |
| [1] | 马利花, 侯梦娟, 朱新霞. 陆地棉GhNFD4在棉花干旱响应中的功能[J]. 生物技术通报, 2025, 41(3): 104-111. |
| [2] | 丁若羲, 豆硕, 安叶芝, 孔文慧, 郭文静, 张冬梅, 王省芬, 马峙英, 吴立柱. VIGS技术在棉花全生长周期中的应用拓展研究[J]. 生物技术通报, 2025, 41(2): 58-64. |
| [3] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [4] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| [5] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [6] | 展艳, 周利斌, 金文杰, 杜艳, 余丽霞, 曲颖, 马永贵, 刘瑞媛. 辐射诱导植物叶色突变的研究进展[J]. 生物技术通报, 2023, 39(8): 106-113. |
| [7] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [8] | 邓嘉辉, 雷建峰, 赵燚, 刘敏, 胡子曜, 尤扬子, 邵武奎, 柳建飞, 刘晓东. 基于Csy4与MCP的新型迷你基因组编辑系统的构建[J]. 生物技术通报, 2023, 39(10): 68-79. |
| [9] | 朱金成, 杨洋, 娄慧, 张薇. 外源褪黑素调控棉花枯萎病抗性研究[J]. 生物技术通报, 2023, 39(1): 243-252. |
| [10] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| [11] | 赵曾强, 郭文婷, 张析, 李潇玲, 张薇. 棉花抗枯萎病相关基因GhERF5-4D的克隆及功能分析[J]. 生物技术通报, 2022, 38(4): 193-201. |
| [12] | 王娅丽, 周利利, 王娜, 程红梅. 利用流式细胞仪快速鉴定棉花倍性的方法比较[J]. 生物技术通报, 2022, 38(12): 144-148. |
| [13] | 胡华冉, 杜磊, 张芮豪, 钟秋月, 刘发万, 桂敏. 辣椒适应非生物胁迫的研究进展[J]. 生物技术通报, 2022, 38(12): 58-72. |
| [14] | 赵燚, 雷建峰, 刘敏, 胡子曜, 代培红, 刘超, 李月, 刘晓东. CLCrV介导的VIGE体系承载力的研究[J]. 生物技术通报, 2022, 38(11): 210-219. |
| [15] | 胡子曜, 代培红, 刘超, 玛迪娜·木拉提, 王倩, 吾尕力汗·阿不都维力, 赵燚, 孙玲, 徐诗佳, 李月. 陆地棉小GTP结合蛋白基因GhROP3的克隆、表达及VIGS载体的构建[J]. 生物技术通报, 2021, 37(9): 106-113. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||