








生物技术通报 ›› 2025, Vol. 41 ›› Issue (10): 321-333.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0314
黄楚蓝1( ), 曾睿2, 陈佩榕2, 赵亚荣2(
), 曾睿2, 陈佩榕2, 赵亚荣2( ), 王旭2, 姚冬生1(
), 王旭2, 姚冬生1( )
)
收稿日期:2025-03-26
出版日期:2025-10-26
发布日期:2025-10-28
通讯作者:
赵亚荣,女,博士,助理研究员,研究方向 :农产品质量安全;E-mail: zyr520zyr@163.com;作者简介:黄楚蓝,女,硕士研究生,研究方向 :生物化学与分子生物学;E-mail: 1515817278@qq.com
基金资助:
HUANG Chu-lan1( ), ZENG Rui2, CHEN Pei-rong2, ZHAO Ya-rong2(
), ZENG Rui2, CHEN Pei-rong2, ZHAO Ya-rong2( ), WANG Xu2, YAO Dong-sheng1(
), WANG Xu2, YAO Dong-sheng1( )
)
Received:2025-03-26
Published:2025-10-26
Online:2025-10-28
摘要:
目的 探究乙酰羟酸合成酶(AHAS)在黄曲霉毒素合成中的作用机制,特别是其通过代谢网络调控黄曲霉毒素合成的分子机理。 方法 采用4D Label-free蛋白质组学技术,系统地比较ΔAflaILVB/G/I菌株与野生型菌株的蛋白质表达谱差异,对获得的差异蛋白进行GO富集、KEGG富集和蛋白互作等分析。 结果 质谱分析结果显示,共鉴定出1 158个差异表达蛋白,其中521个表达上调,637个表达下调,与黄曲霉毒素合成相关的17个差异蛋白中,有14个明确属于黄曲霉毒素合成基因簇上的蛋白且均呈现显著性表达下调,该结果与RNA-Seq和RT-qPCR结果相对一致。将这17个蛋白与支链氨基酸合成相关的9个差异蛋白互作网络分析发现,AHAS对黄曲霉毒素合成的调控机制可能不是通过关键节点蛋白来实现,而是通过影响代谢通路的物质分配来实现的。 结论 ΔAflaILVB/G/I菌株通过干扰缬氨酸和异亮氨酸的生物合成导致琥珀酰辅酶A和琥珀酸生成不足,进而引发一系列代谢重编程。这种代谢重编程不仅抑制黄曲霉的生长,还影响黄曲霉毒素的合成。
黄楚蓝, 曾睿, 陈佩榕, 赵亚荣, 王旭, 姚冬生. 基于4D Label-free技术研究AflaILVB/G/I基因对黄曲霉毒素合成的蛋白质组学分析[J]. 生物技术通报, 2025, 41(10): 321-333.
HUANG Chu-lan, ZENG Rui, CHEN Pei-rong, ZHAO Ya-rong, WANG Xu, YAO Dong-sheng. Proteomic Analysis Reveals the Role of AflaILVB/G/Ⅰ Gene in Aflatoxin Biosynthesis Based on 4D Label-free Technology[J]. Biotechnology Bulletin, 2025, 41(10): 321-333.
菌株名称 Name of strain | 基因型 Genotype | 来源 Source |
|---|---|---|
| A. flavus NRRL 3357 TJES20.1 | Δku70,ΔargB:: AfpyrG (Wild-type) | 江苏师范大学杨教授惠赠 |
| ΔAflaILVB/G/I(AFLA_000930) | Δku70,ΔargB:: AfpyrG,ΔAflaILVB/G/I:: argB | 本研究构建 |
表1 本研究用到的曲霉菌株
Table 1 Aspergillus strains used in this study
菌株名称 Name of strain | 基因型 Genotype | 来源 Source |
|---|---|---|
| A. flavus NRRL 3357 TJES20.1 | Δku70,ΔargB:: AfpyrG (Wild-type) | 江苏师范大学杨教授惠赠 |
| ΔAflaILVB/G/I(AFLA_000930) | Δku70,ΔargB:: AfpyrG,ΔAflaILVB/G/I:: argB | 本研究构建 |
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Actin-QF | ACGGTGTCGTCACAAACTGG |
| Actin-QR | CGGTTGGACTTAGGGTTGATAG |
| aflP-QF | ACGAAGCCACTGGTAGAGGAGATG |
| aflP-QR | GTGAATGACGGCAGGCAGGT |
| aflO-QF | GATTGGGATGTGGTCATGCGATT |
| aflO-QR | GCCTGGGTCCGAAGAATGC |
| aflK-QF | CTCGCACTTTGGCATGTACG |
| aflK-QR | AATCCTCCCGCCTCAATCAC |
| aflD-QF | GTGGTGGTTGCCAATGCG |
| aflD-QR | CTGAAACAGTAGGACGGGAGC |
| hypC-QF | GCATGGTGCCTTACACATGG |
| hypC-QR | CCTACCAACCTCACGCTCTC |
表2 荧光定量引物序列
Table 2 Primer sequences for fluorescence quantification
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Actin-QF | ACGGTGTCGTCACAAACTGG |
| Actin-QR | CGGTTGGACTTAGGGTTGATAG |
| aflP-QF | ACGAAGCCACTGGTAGAGGAGATG |
| aflP-QR | GTGAATGACGGCAGGCAGGT |
| aflO-QF | GATTGGGATGTGGTCATGCGATT |
| aflO-QR | GCCTGGGTCCGAAGAATGC |
| aflK-QF | CTCGCACTTTGGCATGTACG |
| aflK-QR | AATCCTCCCGCCTCAATCAC |
| aflD-QF | GTGGTGGTTGCCAATGCG |
| aflD-QR | CTGAAACAGTAGGACGGGAGC |
| hypC-QF | GCATGGTGCCTTACACATGG |
| hypC-QR | CCTACCAACCTCACGCTCTC |

图1 质谱数据质量控制A:蛋白质谱总鉴定; B:特异性肽段的电荷和长度分布; C:定量蛋白的肽段数量分布; D:定量蛋白的相对分子质量分布
Fig. 1 Quality control of mass spectrometry dataA: Total protein identification by mass spectrometry. B: Charge and length distribution of specific peptides. C: Peptide number distribution of quantified proteins. D: Relative molecular weight distribution of quantified proteins
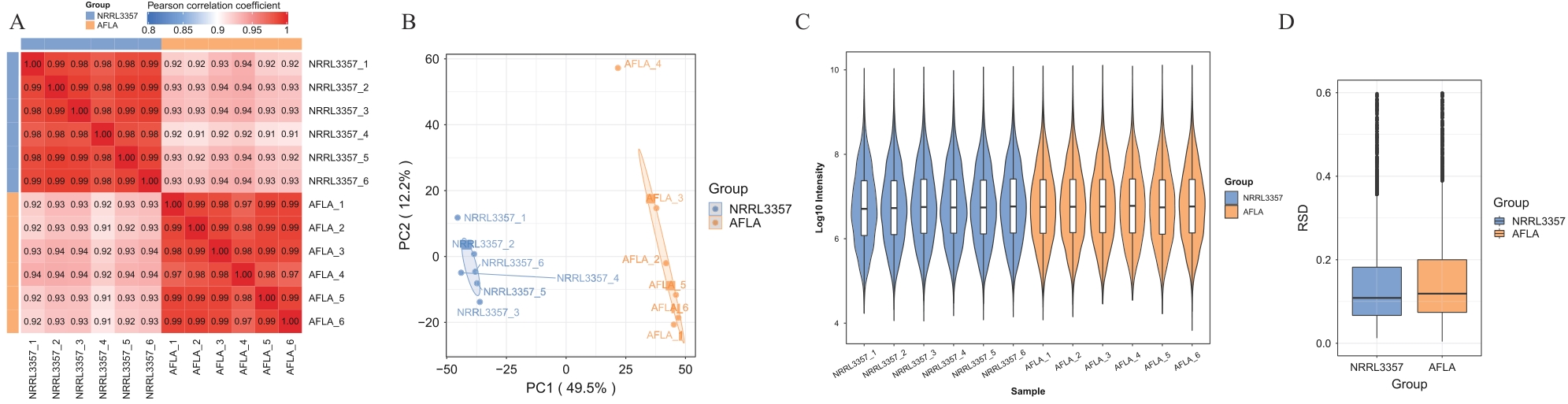
图2 样本重复性分析A:PCC分析热图; B:PCA主成分分析; C:RSD分析小提琴图; D:RSD分析箱线图
Fig. 2 Sample reproducibility analysisA: PCC analysis heatmap. B: PCA Principal component analysis. C:RSD Analysis violin plot. D: RSD analysis boxplot

图4 差异蛋白GO功能富集分析A:上调的差异蛋白GO富集气泡图; B:下调的差异蛋白GO富集气泡图
Fig. 4 GO functional enrichment analysis of differentially expressed proteinsA: GO enrichment bubble plot of upregulated differentially expressed proteins. B: GO enrichment bubble plot of downregulated differentially expressed proteins

图5 差异蛋白KEGG功能富集分析A:上调的差异蛋白KEGG富集条形图; B:下调的差异蛋白KEGG富集条形图
Fig. 5 KEGG functional enrichment analysis of differentially expressed proteinsA: KEGG enrichment bar plot of upregulated differentially expressed proteins. B: KEGG enrichment bar plot of downregulated differentially expressed protein
蛋白质编码 Protein accession | 蛋白质描述 Protein description | 基因名称 Gene name | P值 P value | 调控类型Regulated type |
|---|---|---|---|---|
| B8MYP9 | Branched-chain amino acid aminotransferase | AFLA_081880 | 2.204 49E-08 | Down |
| B8N7W8 | Dihydroxy acid dehydratase Ilv3, putative | AFLA_105610 | 4.045 78E-08 | Up |
| B8N9W8 | Acetohydroxy-acid synthase small subunit | AFLA_112640 | 4.260 63E-08 | Up |
| B8N9L9 | Threonine dehydratase | AFLA_111650 | 9.742 79E-08 | Up |
| B8NEC1 | Ketol-acid reductoisomerase, mitochondrial | AFLA_060300 | 1.855 62E-06 | Up |
| B8N1U6 | Pyruvate decarboxylase | AFLA_032500 | 2.141 48E-06 | Up |
| B8N9Q5 | L-serine dehydratase, putative | AFLA_112010 | 7.377 42E-05 | Down |
| B8NEE4 | Pyridoxal-phosphate dependent enzyme, putative | AFLA_060530 | 0.000 130 39 | Down |
| B8NB44 | Branched-chain-amino-acid aminotransferase | AFLA_044190 | 0.000 135 251 | Up |
表3 支链氨基酸合成途径涉及的差异蛋白
Table 3 Differentially expressed proteins involved in the branched-chain amino acid biosynthesis pathway
蛋白质编码 Protein accession | 蛋白质描述 Protein description | 基因名称 Gene name | P值 P value | 调控类型Regulated type |
|---|---|---|---|---|
| B8MYP9 | Branched-chain amino acid aminotransferase | AFLA_081880 | 2.204 49E-08 | Down |
| B8N7W8 | Dihydroxy acid dehydratase Ilv3, putative | AFLA_105610 | 4.045 78E-08 | Up |
| B8N9W8 | Acetohydroxy-acid synthase small subunit | AFLA_112640 | 4.260 63E-08 | Up |
| B8N9L9 | Threonine dehydratase | AFLA_111650 | 9.742 79E-08 | Up |
| B8NEC1 | Ketol-acid reductoisomerase, mitochondrial | AFLA_060300 | 1.855 62E-06 | Up |
| B8N1U6 | Pyruvate decarboxylase | AFLA_032500 | 2.141 48E-06 | Up |
| B8N9Q5 | L-serine dehydratase, putative | AFLA_112010 | 7.377 42E-05 | Down |
| B8NEE4 | Pyridoxal-phosphate dependent enzyme, putative | AFLA_060530 | 0.000 130 39 | Down |
| B8NB44 | Branched-chain-amino-acid aminotransferase | AFLA_044190 | 0.000 135 251 | Up |
蛋白质编码 Protein accession | 蛋白质描述 Protein description | 基因名称 Gene name | P值 P value | 调控类型Regulated type |
|---|---|---|---|---|
| P55790 | Sterigmatocystin 8-O-methyltransferase | omtA/aflP/AFLA_139210 | 1.205 77E-13 | Down |
| Q9P900 | Demethylsterigmatocystin 6-O-methyltransferase | omtB/aflO/AFLA_139220 | 1.503 66E-13 | Down |
| B8NHY3 | AflK/VERB synthase | vbs/aflK/AFLA_139190 | 1.187 86E-12 | Down |
| B8NHZ4 | AflM/dehydrogenase/ketoreductase | ver-1/aflM/AFLA_139300 | 6.337 76E-12 | Down |
| B8NI02 | AflD/reductase | nor-1/aflD/AFLA_139390 | 1.591 52E-11 | Down |
| B8NHY1 | AflW/monooxygenase | moxY/aflW/AFLA_139170 | 2.287 05E-11 | Down |
| B8NHX9 | AflY | hypA/aflY/AFLA_139150 | 3.178 44E-11 | Down |
| B8N9Y6 | Toxin biosynthesis ketoreductase | AFLA_112820 | 3.624 58E-11 | Up |
| B8NHY0 | AflX/monooxygenase/oxidase | ordB/aflX/AFLA_139160 | 2.447 17E-09 | Down |
| B8NI03 | Noranthrone monooxygenase | hypC/AFLA_139400 | 6.130 27E-09 | Down |
| B8NT69 | O-methyltransferase | AFLA_053520 | 3.688 78E-08 | Down |
| B8NHY9 | AflL/desaturase/ P450 monooxygenase | verb/aflL/AFLA_139250 | 5.666 24E-08 | Down |
| B8NHZ6 | AflJ/esterase | estA/aflJ/AFLA_139320 | 8.454 44E-08 | Down |
| B8NHZ2 | AflN/monooxygenase | ver-A/aflN/AFLA_139280 | 9.288 83E-08 | Down |
| B8NI04 | AflC/polyketide synthase | pskA/aflC/AFLA_139410 | 1.609 73E-05 | Down |
| B8N0Z3 | N-alkane-inducible cytochrome P450 | AFLA_027680 | 0.000 725 794 | Up |
| B8N628 | O-methyltransferase family protein | AFLA_016120 | 0.017 023 299 | Down |
表4 黄曲霉毒素合成途径涉及的差异蛋白
Table 4 Differentially expressed proteins involved in the aflatoxin biosynthesis pathway
蛋白质编码 Protein accession | 蛋白质描述 Protein description | 基因名称 Gene name | P值 P value | 调控类型Regulated type |
|---|---|---|---|---|
| P55790 | Sterigmatocystin 8-O-methyltransferase | omtA/aflP/AFLA_139210 | 1.205 77E-13 | Down |
| Q9P900 | Demethylsterigmatocystin 6-O-methyltransferase | omtB/aflO/AFLA_139220 | 1.503 66E-13 | Down |
| B8NHY3 | AflK/VERB synthase | vbs/aflK/AFLA_139190 | 1.187 86E-12 | Down |
| B8NHZ4 | AflM/dehydrogenase/ketoreductase | ver-1/aflM/AFLA_139300 | 6.337 76E-12 | Down |
| B8NI02 | AflD/reductase | nor-1/aflD/AFLA_139390 | 1.591 52E-11 | Down |
| B8NHY1 | AflW/monooxygenase | moxY/aflW/AFLA_139170 | 2.287 05E-11 | Down |
| B8NHX9 | AflY | hypA/aflY/AFLA_139150 | 3.178 44E-11 | Down |
| B8N9Y6 | Toxin biosynthesis ketoreductase | AFLA_112820 | 3.624 58E-11 | Up |
| B8NHY0 | AflX/monooxygenase/oxidase | ordB/aflX/AFLA_139160 | 2.447 17E-09 | Down |
| B8NI03 | Noranthrone monooxygenase | hypC/AFLA_139400 | 6.130 27E-09 | Down |
| B8NT69 | O-methyltransferase | AFLA_053520 | 3.688 78E-08 | Down |
| B8NHY9 | AflL/desaturase/ P450 monooxygenase | verb/aflL/AFLA_139250 | 5.666 24E-08 | Down |
| B8NHZ6 | AflJ/esterase | estA/aflJ/AFLA_139320 | 8.454 44E-08 | Down |
| B8NHZ2 | AflN/monooxygenase | ver-A/aflN/AFLA_139280 | 9.288 83E-08 | Down |
| B8NI04 | AflC/polyketide synthase | pskA/aflC/AFLA_139410 | 1.609 73E-05 | Down |
| B8N0Z3 | N-alkane-inducible cytochrome P450 | AFLA_027680 | 0.000 725 794 | Up |
| B8N628 | O-methyltransferase family protein | AFLA_016120 | 0.017 023 299 | Down |

图8 差异蛋白互作网络分析椭圆形和正方形属于支链氨基酸合成途径差异蛋白互作到蛋白质,其中椭圆形与AFLA_000930蛋白有互作关系;三角形属于黄曲霉毒素合成途径差异蛋白互作到蛋白质;菱形为连接节点蛋白。红色代表上调的差异蛋白;蓝色代表下调的差异蛋白
Fig. 8 Protein-protein interaction network analysis of differentially expressed proteinsEllipses and squares indicate differentially expressed proteins in the branched-chain amino acid synthesis pathway that interact with proteins, where ellipses interact with the AFLA_000930 protein; triangles indicate differentially expressed proteins in the aflatoxin synthesis pathway that interact with proteins; diamonds indicate connecting node proteins. Red indicates upregulated differentially expressed proteins; blue indicates downregulated differentially expressed proteins

图10 细胞内代谢重编程分析×代表该途径受到AHAS功能缺陷干扰;红色代表上调的代谢途径;蓝色代表下调的代谢途径
Fig. 10 Analysis of intracellular metabolic reprogramming× indicates metabolic pathways affected by AHAS functional deficiency; red indicates upregulated metabolic pathways; blue indicates downregulated metabolic pathways
| [1] | 王晓燕, 梁柳柯, 魏闪, 等. AflbasR基因对储粮霉菌黄曲霉生长发育和毒素合成的影响 [J]. 河南工业大学学报: 自然科学版, 2024, 45(6): 45-53. |
| Wang XY, Liang LK, Wei S, et al. Effects of AflbasR gene on the growth, development and aflatoxin biosynthesis of the grain storage fungus Aspergillus flavus [J]. J Henan Univ Technol Nat Sci | |
| Ed, 2024, 45(6): 45-53. | |
| [2] | Pickova D, Ostry V, Malir F. A recent overview of producers and important dietary sources of aflatoxins [J]. Toxins, 2021, 13(3): 186. |
| [3] | 尚艳娥, 杨卫民. CAC、欧盟、美国与中国粮食中真菌毒素限量标准的差异分析 [J]. 食品科学技术学报, 2019, 37(1): 10-15. |
| Shang YE, Yang WM. Variation analysis of cereals mycotoxin limit standards of CAC, EU, USA, and China [J]. J Food Sci Technol, 2019, 37(1): 10-15. | |
| [4] | International Agency for Research on Cancer (IARC), World Health Organization (WHO). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Volume 56: Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins [M]. New York: Cancer causes & control, 1994, 5(1): 89-90. |
| [5] | Xue MY, Qu Z, Moretti A, et al. Aspergillus mycotoxins: the major food contaminants [J]. Adv Sci, 2025, 12(9): e2412757. |
| [6] | Mitchell NJ, Bowers E, Hurburgh C, et al. Potential economic losses to the US corn industry from aflatoxin contamination [J]. Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2016, 33(3): 540-550. |
| [7] | Cheng XB, Vella A, Stasiewicz MJ. Classification of aflatoxin contaminated single corn kernels by ultraviolet to near infrared spectroscopy [J]. Food Control, 2019, 98: 253-261. |
| [8] | Chhaya RS, O’Brien J, Nag R, et al. Prevalence and concentration of mycotoxins in bovine feed and feed components: a global systematic review and meta-analysis [J]. Sci Total Environ, 2024, 929: 172323. |
| [9] | Gruber-Dorninger C, Müller A, Rosen R. Multi-mycotoxin contamination of aquaculture feed: a global survey [J]. Toxins, 2025, 17(3): 116. |
| [10] | 雷元培, 周建川,郑文革, 等. 2019-2020年中国饲料原料和饲料中霉菌毒素污染调查报告 [J]. 饲料工业, 2022, 43(20): 59-64. |
| Lei YP, Zhou JC, Zheng WG, et al. Investigation report on mycotoxin contamination in feed ingredients and feeds in China from 2019 to 2020 [J]. Feed Industry, 2022, 43(20): 59-64. | |
| [11] | Zentai A, Jóźwiak Á, Süth M, et al. Carry-over of aflatoxin B1 from feed to cow milk-a review [J]. Toxins, 2023, 15(3): 195. |
| [12] | Marchese S, Polo A, Ariano A, et al. Aflatoxin B1 and M1: biological properties and their involvement in cancer development [J]. Toxins, 2018, 10(6): 214. |
| [13] | De Baere S, Ochieng PE, Kemboi DC, et al. Development of high-throughput sample preparation procedures for the quantitative determination of aflatoxins in biological matrices of chickens and cattle using UHPLC-MS/MS [J]. Toxins, 2023, 15(1): 37. |
| [14] | Khalil HMA, Eid WAM, El-Nablaway M, et al. Date seeds powder alleviate the aflatoxin B1 provoked heart toxicity in male offspring rat [J]. Sci Rep, 2024, 14(1): 30480. |
| [15] | Cao HH, Molina S, Sumner S, et al. An untargeted metabolomic analysis of acute AFB1 treatment in liver, breast, and lung cells [J]. PLoS One, 2025, 20(1): e0313159. |
| [16] | Herzallah. Aflatoxin b1 residues in eggs and flesh of laying hens fed aflatoxin b1 contaminated diet [J]. Am J Agric Biol Sci, 2013, 8(2): 156-161. |
| [17] | He XN, Zeng ZZ, Jiang WD, et al. Aflatoxin B1 decreased flesh flavor and inhibited muscle development in grass carp (Ctenopharyngodon idella) [J]. Anim Nutr, 2024, 18: 27-38. |
| [18] | Liu H, Xie RT, Huang WB, et al. Effects of dietary aflatoxin B1 on hybrid grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) growth, intestinal health, and muscle quality [J]. Aquac Nutr, 2024, 2024: 3920254. |
| [19] | Pożarska A, Karpiesiuk K, Kozera W, et al. AFB1 toxicity in human food and animal feed consumption: a review of experimental treatments and preventive measures [J]. Int J Mol Sci, 2024, 25(10): 5305. |
| [20] | Benkerroum N, Ismail A. Human breast milk contamination with aflatoxins, impact on children’s health, and possible control means: a review [J]. Int J Environ Res Public Health, 2022, 19(24): 16792. |
| [21] | Liang YF, Long ZX, Zhang YJ, et al. The chemical mechanisms of the enzymes in the branched-chain amino acids biosynthetic pathway and their applications [J]. Biochimie, 2021, 184: 72-87. |
| [22] | Lonhienne T, Low YS, Garcia MD, et al. Structures of fungal and plant acetohydroxyacid synthases [J]. Nature, 2020, 586(7828): 317-321. |
| [23] | Liu YD, Li YY, Wang XY. Acetohydroxyacid synthases: evolution, structure, and function [J]. Appl Microbiol Biotechnol, 2016, 100(20): 8633-8649. |
| [24] | Zhang YY, Li Y, Liu X, et al. Molecular architecture of the acetohydroxyacid synthase holoenzyme [J]. Biochem J, 2020, 477(13): 2439-2449. |
| [25] | Chong NF, Van de Wouw AP, Idnurm A. The ilv2 gene, encoding acetolactate synthase for branched chain amino acid biosynthesis, is required for plant pathogenicity by Leptosphaeria maculans [J]. Mol Biol Rep, 2024, 51(1): 682. |
| [26] | Low YS, Garcia MD, Lonhienne T, et al. Triazolopyrimidine herbicides are potent inhibitors of Aspergillus fumigatus acetohydroxyacid synthase and potential antifungal drug leads [J]. Sci Rep, 2021, 11(1): 21055. |
| [27] | Agnew-Francis KA, Tang YC, Lin X, et al. Herbicides that target acetohydroxyacid synthase are potent inhibitors of the growth of drug-resistant Candida auris [J]. ACS Infect Dis, 2020, 6(11): 2901-2912. |
| [28] | Shao SN, Li B, Sun Q, et al. Acetolactate synthases regulatory subunit and catalytic subunit genes VdILVs are involved in BCAA biosynthesis, microscletotial and conidial formation and virulence in Verticillium dahlia e [J]. Fungal Genet Biol, 2022, 159: 103667. |
| [29] | Du Y, Zhang HF, Hong L, et al. Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae [J]. Mol Plant Pathol, 2013, 14(9): 870-884. |
| [30] | Liu X, Han Q, Xu JH, et al. Acetohydroxyacid synthase FgIlv2 and FgIlv6 are involved in BCAA biosynthesis, mycelial and conidial morphogenesis, and full virulence in Fusarium graminearum [J]. Sci Rep, 2015, 5: 16315. |
| [31] | Zhao YR, Huang CL, Zeng R, et al. AflaILVB/G/I and AflaILVD are involved in mycelial production, aflatoxin biosynthesis, and fungal virulence in Aspergillus flavus [J]. Front Cell Infect Microbiol, 2024, 14: 1372779. |
| [32] | Cimbalo A, Frangiamone M, Font G, et al. The importance of transcriptomics and proteomics for studying molecular mechanisms of mycotoxin exposure: a review [J]. Food Chem Toxicol, 2022, 169: 113396. |
| [33] | Lv YY, Lv A, Zhai HC, et al. Insight into the global regulation of laeA in Aspergillus flavus based on proteomic profiling [J]. Int J Food Microbiol, 2018, 284: 11-21. |
| [34] | Zhang F, Zhong H, Han XY, et al. Proteomic profile of Aspergillus flavus in response to water activity [J]. Fungal Biol, 2015, 119(2/3): 114-124. |
| [35] | Bai YH, Wang S, Zhong H, et al. Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature [J]. Sci Rep, 2015, 5: 14582. |
| [36] | Tiwari S, Thakur R, Goel G, et al. Nano-LC-Q-TOF analysis of proteome revealed germination of Aspergillus flavus conidia is accompanied by MAPK signalling and cell wall modulation [J]. Mycopathologia, 2016, 181(11-12): 769-786. |
| [37] | Khan R, Ghazali FM, Mahyudin NA, et al. Aflatoxin biosynthesis, genetic regulation, toxicity, and control strategies: a review [J]. J Fungi, 2021, 7(8): 606. |
| [38] | McCourt JA, Duggleby RG. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids [J]. Amino Acids, 2006, 31(2): 173-210. |
| [39] | Yamamoto K, Tsuchisaka A, Yukawa H. Branched-chain amino acids [M]//Amino Acid Fermentation. Tokyo: Springer Japan, 2016: 103-128. |
| [40] | Nie CX, He T, Zhang WJ, et al. Branched chain amino acids: beyond nutrition metabolism [J]. Int J Mol Sci, 2018, 19(4): 954. |
| [41] | Neinast M, Murashige D, Arany Z. Branched chain amino acids [J]. Annu Rev Physiol, 2019, 81: 139-164. |
| [42] | Sivanand S, Vander Heiden MG. Emerging roles for branched-chain amino acid metabolism in cancer [J]. Cancer Cell, 2020, 37(2): 147-156. |
| [43] | Dimou A, Tsimihodimos V, Bairaktari E. The critical role of the branched chain amino acids (BCAAs) catabolism-regulating enzymes, branched-chain aminotransferase (BCAT) and branched-chain α- keto acid dehydrogenase (BCKD), in human pathophysiology [J]. Int J Mol Sci, 2022, 23(7): 4022. |
| [44] | Huang ZC, Wang Q, Khan IA, et al. The methylcitrate cycle and its crosstalk with the glyoxylate cycle and tricarboxylic acid cycle in pathogenic fungi [J]. Molecules, 2023, 28(18): 6667. |
| [45] | Akram M. Citric acid cycle and role of its intermediates in metabolism [J]. Cell Biochem Biophys, 2014, 68(3): 475-478. |
| [46] | Huang YM, Huber GA, Wang N, et al. Brownian dynamic study of an enzyme metabolon in the TCA cycle: Substrate kinetics and channeling [J]. Protein Sci, 2018, 27(2): 463-471. |
| [47] | Chang LC, Chiang SK, Chen SE, et al. Targeting 2-oxoglutarate dehydrogenase for cancer treatment [J]. Am J Cancer Res, 2022, 12(4): 1436-1455. |
| [48] | Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement [J]. Redox Biol, 2020, 37: 101674. |
| [49] | MacLean A, Legendre F, Appanna VD. The tricarboxylic acid (TCA) cycle: a malleable metabolic network to counter cellular stress [J]. Crit Rev Biochem Mol Biol, 2023, 58(1): 81-97. |
| [50] | Liu XJ, Si WZ, He L, et al. The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry [J]. Signal Transduct Target Ther, 2021, 6(1): 375. |
| [51] | Doan MT, Teitell MA. Krebs and an alternative TCA cycle! [J]. Cell Res, 2022, 32(6): 509-510. |
| [52] | Arnold PK, Jackson BT, Paras KI, et al. A non-canonical tricarboxylic acid cycle underlies cellular identity [J]. Nature, 2022, 603(7901): 477-481. |
| [53] | Yan SJ, Liang YT, Zhang JD, et al. Aspergillus flavus grown in peptone as the carbon source exhibits spore density- and peptone concentration-dependent aflatoxin biosynthesis [J]. BMC Microbiol, 2012, 12: 106. |
| [1] | 蒋天威, 马培杰, 李亚娇, 陈才俊, 刘晓霞, 王小利. 二穗短柄草对光周期的代谢响应分析[J]. 生物技术通报, 2025, 41(7): 237-247. |
| [2] | 王碧成, 景海青, 万坤, 张莹莹, 丁家豪, 李润植, 薛金爱, 张海平. 大豆BCAT基因家族鉴定及GmBCAT3在干旱胁迫中的功能分析[J]. 生物技术通报, 2025, 41(10): 196-209. |
| [3] | 杨微, 关海峰, 任欣慧, 彭金菊, 陈志宝. 虾青素对黄曲霉毒素B1诱导肝损伤的缓解作用及机制[J]. 生物技术通报, 2025, 41(10): 334-342. |
| [4] | 杨冬, 唐璎. 枯草芽孢杆菌WTX1胞外酶降解AFB1酶学特性及降解位点分析[J]. 生物技术通报, 2023, 39(4): 93-102. |
| [5] | 赵雅茹, 许庆方, 高文俊, 郭刚, 陈雷, 玉柱. 抑霉乳酸菌脱毒特性及青贮应用的研究[J]. 生物技术通报, 2021, 37(9): 95-105. |
| [6] | 唐璎, 黄佳, 邓展瑞, 杨晓楠. 一株枯草芽孢杆菌降解黄曲霉毒素B1产物分析[J]. 生物技术通报, 2021, 37(12): 82-90. |
| [7] | 刘端木, 吴怿, 刘沄, 梁志宏. 一株产毒曲霉拮抗细菌的筛选、鉴定及抑菌活性研究[J]. 生物技术通报, 2019, 35(8): 42-50. |
| [8] | 耿龙泼, 王鑫旺, 黄路华, 邓基利, 汪世华, 张峰. 黄曲霉核糖体蛋白基因在不同生长时期的表达分析[J]. 生物技术通报, 2018, 34(4): 194-200. |
| [9] | 任亚林, 李耘, 吴静. AFB1和ZEN对HepG2细胞体外联合毒性效应及机制研究[J]. 生物技术通报, 2018, 34(11): 160-167. |
| [10] | 曾昆,杜道林,薛永来. 基于特异性生物识别分子的黄曲霉毒素快速分析方法研究进展[J]. 生物技术通报, 2016, 32(8): 47-55. |
| [11] | 庞志伟, 路旭, 胡江春 , 陈晓琦, 王楠, 宋艳玲. 诱导烟草悬浮细胞氧爆发的海绵共附生真菌HMP-F66的筛选与鉴定[J]. 生物技术通报, 2015, 31(7): 174-179. |
| [12] | 路子显;伍松陵;孙长坡;. 黄曲霉素合成相关基因表达与环境因素的关系[J]. , 2010, 0(11): 56-61. |
| [13] | 李科;孙彦平;蔡小波;赵海;. 黄曲霉产木聚糖酶条件优化及酶解产物初步分析[J]. , 2009, 0(S1): 348-351. |
| [14] | 朱遐;. 农业诊断剂市场[J]. , 1993, 0(01): 7-8. |
| [15] | 金人一;. 美Idexx公司出售化验牛奶的新技术[J]. , 1989, 0(09): 32-32. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||