








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 114-123.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0404
收稿日期:2025-04-17
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
孟超敏,男,副教授,研究方向 :旱地特色作物遗传育种与栽培;E-mail: chaominm@haust.edu.cn作者简介:郭浩杰,男,硕士研究生,研究方向 :谷子遗传育种;E-mail: 2214442069@qq.com
基金资助:
GUO Hao-jie( ), WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min(
), WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min( )
)
Received:2025-04-17
Published:2025-12-26
Online:2026-01-06
摘要:
目的 SPX基因家族在植物磷的信号感知、吸收和运输中发挥着重要的功能。探究SiSPX9在响应低磷胁迫过程中的功能,为解析谷子耐低磷机制提供理论依据。 方法 从谷子根中克隆SiSPX9,并对其进行生物信息学分析。利用实时荧光定量PCR(real-time quantitative PCR, RT-qPCR)分析SiSPX9在不同组织中的表达及不同低磷胁迫时长处理的表达模式。通过农杆菌介导的遗传转化法在拟南芥中异源表达SiSPX9,并对转基因植株进行低磷处理下发芽率、根长、根系表面积的检测。 结果 SiSPX9基因CDS全长759 bp,编码253个氨基酸,具有SPX家族特征保守结构域;蛋白质结构预测结果显示,其编码的蛋白质二级结构由68.38% α-螺旋、1.19% β-折叠、30.43%的延伸链构成,三级结构与二级结构相统一;通过同源进化分析发现,谷子SiSPX9与高粱和玉米相似性最高,在进化上属于同一分支。RT-qPCR分析结果表明,SiSPX9基因在各个组织的表达特征存在差异性,在根中表达量最高,在叶中表达量最低;在不同低磷胁迫时长处理下,SiSPX9随着低磷胁迫时间的增加表达量呈先上升后下降的趋势,在低磷胁迫12 h时其表达量达到最高值,是适磷处理的15.3倍。功能验证分析结果表明,在低磷胁迫下,野生型(Col-0)和SiSPX9过表达拟南芥株系的萌发率、根长和根系表面积均受到一定程度抑制,但SiSPX9过表达株系明显优于Col-0。 结论 SiSPX9过表达显著增强拟南芥在低磷胁迫下的萌发率和根系发育,表明其通过调控磷信号通路提升耐低磷胁迫的能力。
郭浩杰, 王成, 杨馥熔, 杜冰, 孟超敏. 谷子SiSPX9基因的克隆及耐低磷分析[J]. 生物技术通报, 2025, 41(12): 114-123.
GUO Hao-jie, WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min. Cloning and Low-phosphorus Tolerance Analysis of SiSPX9 Gene in Foxtail Millet[J]. Biotechnology Bulletin, 2025, 41(12): 114-123.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| SiSPX9-F | ATGAAGTTCGGCAAGCGG | SiSPX9 CDS扩增 |
| SiSPX9-R | TCACTGCGTGGGGATGAG | |
| SiSPX9-qF | TTCGTGGACTACTGCCTGCTC | SiSPX9 RT-qPCR 分析 |
| SiSPX9-qR | GCTTGTCTGCTGCTGCTGTG | |
| Actin-qF | GGCAAACAGGGAGAAGATGA | 谷子内参基因 RT-qPCR 分析 |
| Actin-qR | GAGGTTGTCGGTAAGGTCACG | |
| SiSPX9-pBI121-F | CGCGGATCCATGAAGTTCGGCAAGCGG | pBI121-SiSPX9 载体构建 |
| SiSPX9-pBI121-R | CGCGAGCTCTCACTGCGTGGGGATGAG | |
| 35S-R | GAGCTCCTTTTCCAGCGGACC | 转基因拟南芥分子鉴定 |
表1 本试验用到的引物
Table 1 Primers used in this experiment
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| SiSPX9-F | ATGAAGTTCGGCAAGCGG | SiSPX9 CDS扩增 |
| SiSPX9-R | TCACTGCGTGGGGATGAG | |
| SiSPX9-qF | TTCGTGGACTACTGCCTGCTC | SiSPX9 RT-qPCR 分析 |
| SiSPX9-qR | GCTTGTCTGCTGCTGCTGTG | |
| Actin-qF | GGCAAACAGGGAGAAGATGA | 谷子内参基因 RT-qPCR 分析 |
| Actin-qR | GAGGTTGTCGGTAAGGTCACG | |
| SiSPX9-pBI121-F | CGCGGATCCATGAAGTTCGGCAAGCGG | pBI121-SiSPX9 载体构建 |
| SiSPX9-pBI121-R | CGCGAGCTCTCACTGCGTGGGGATGAG | |
| 35S-R | GAGCTCCTTTTCCAGCGGACC | 转基因拟南芥分子鉴定 |
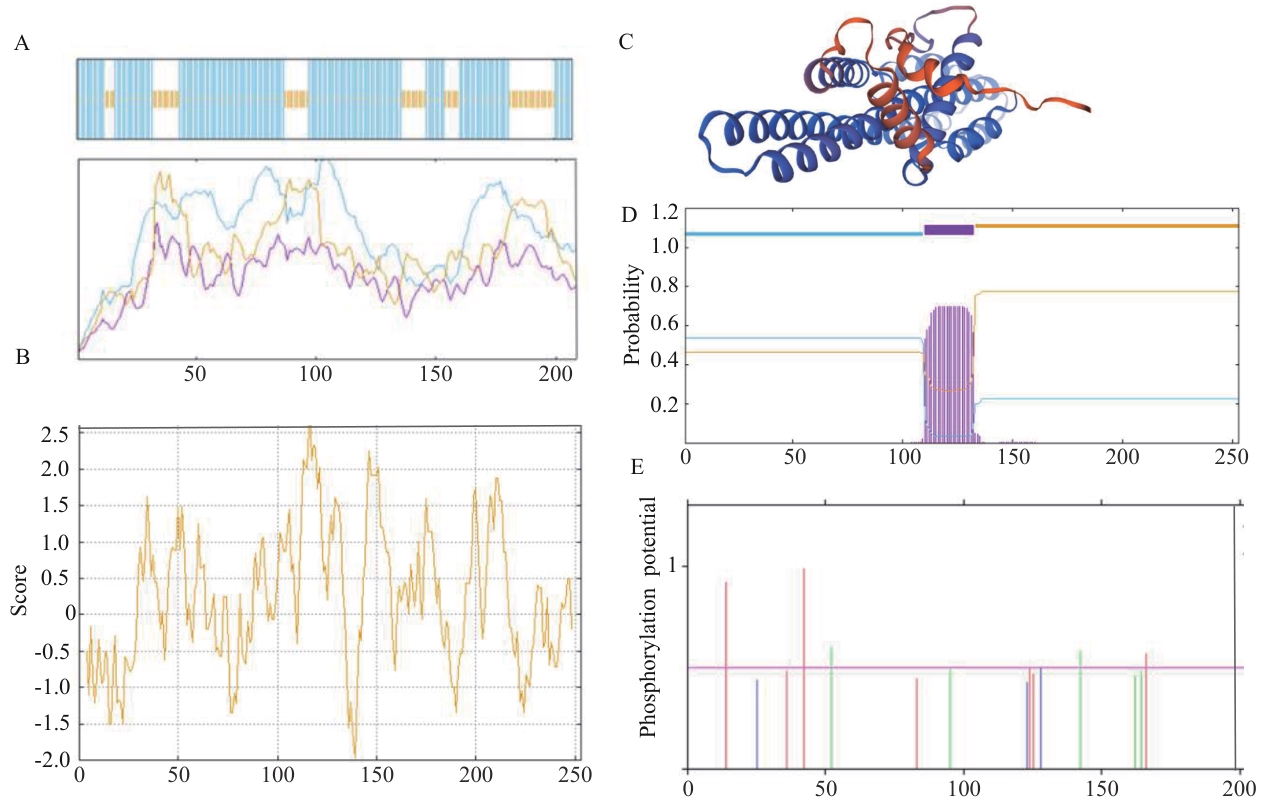
图3 SiSPX9蛋白的生物信息学分析A:二级结构分析;B:三级结构预测;C:蛋白质亲/疏水性预测;D:蛋白跨膜结构预测;E:磷酸化位点预测
Fig. 3 Bioinformatics analysis ofSiSPX9proteinA: Prediction of secondary structure. B: Tertiary structure of SiSPX9 protein. C: Protein hydrophilicity/hydrophobicity prediction. D: Protein transmembrane structure prediction. E: Phosphorylation site prediction
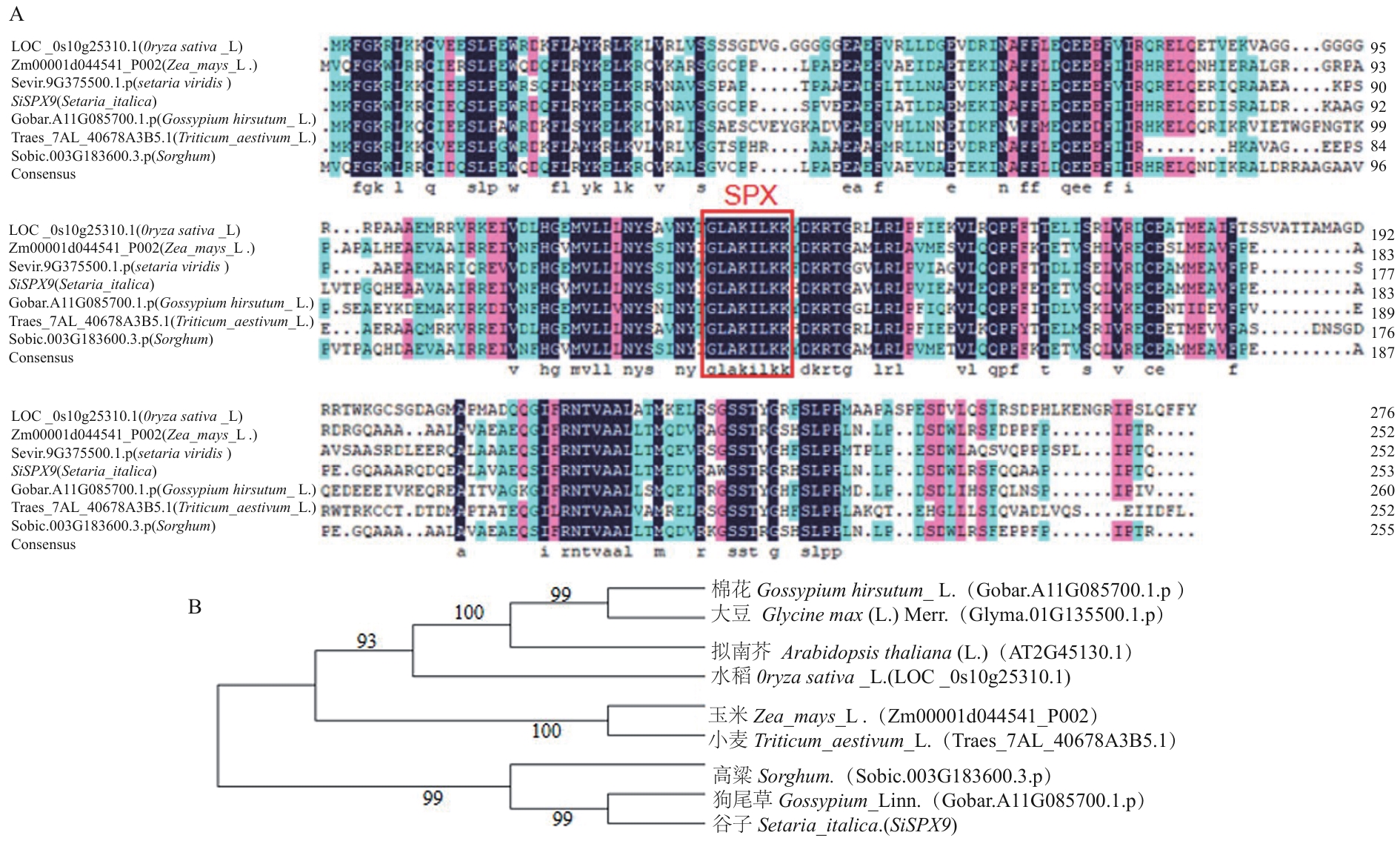
图4 SiSPX9蛋白序列比对和进化树构建A:不同植物SiSPX9蛋白序列比对,图中红色方框表示SPX蛋白的结构域;B:SiSPX9基因系统进化树
Fig. 4 SiSPX9 protein sequence alignment and phylogenetic tree constructionA: Alignment of SiSPX9protein sequences from different plants. The red box in the diagram contains the domains ofSPX protein. B: Phylogenetic tree analysis of SiSPX9 gene
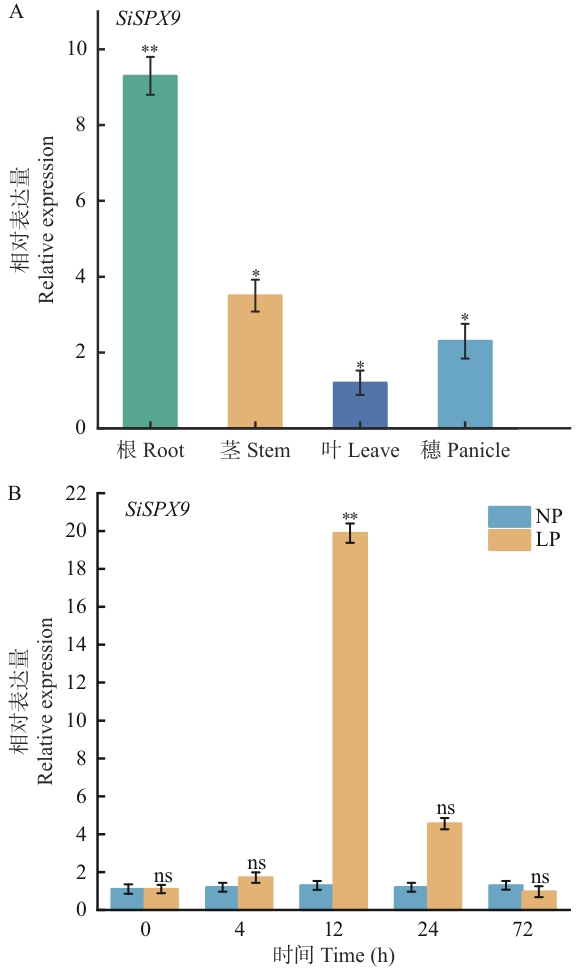
图5 谷子不同组织及低磷胁迫下SiSPX9的表达分析A:SiSPX9在谷子不同组织中的表达;B:SiSPX9在不同低磷胁迫时间下根中的表达情况,NP:正常磷处理(1 mmol/L KH₂PO₄);LP:低磷处理(1 μmol/L KH₂PO₄);*P<0.05,**P<0.01,ns表示差异不显著(P>0.05),下同
Fig. 5 Tissue-specific and low-phosphorus stress-induced expression of SiSPX9 in foxtail milletA: Expressions of SiSPX9 in different tissues of foxtail millet (Setaria italica). B: Expression patterns of SiSPX9 in roots under low-phosphorus stress at different time points. NP: Normal phosphorus treatment (1 mmol/L KH₂PO₄); LP: Low phosphorus treatment (1 μmol/L KH₂PO₄). *P<0.05, **P< 0.01, ns indicates no significant difference (P>0.05). The same below
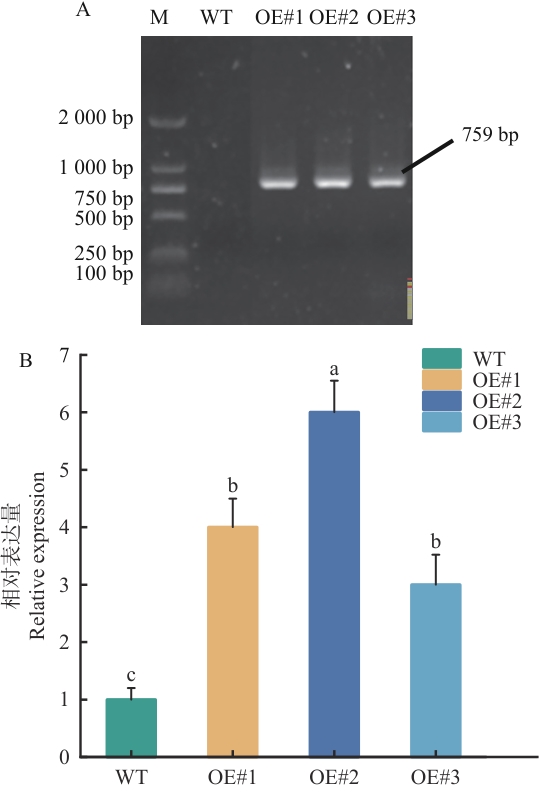
图6 转基因SiSPX9拟南芥鉴定结果及表达量分析A:转基因拟南芥PCR检测电泳图(M:DL 2000 DNA marker;WT:野生型拟南芥;OE#1-OE1#3:转pBI121-SiSPX9拟南芥);B:转基因拟南芥qPCR检测,OE#1-OE#3代表过表达阳性植株;显著性差异通过Duncan新复极差法确定;不同字母表示差异显著(P<0.05);误差线表示3个独立生物学实验获得的标准差;以Actin为内参对目的基因相对表达量进行标准化计算。下同
Fig. 6 Identificatied results of transgenic SiSPX9A. thaliana andexpression analysisA: PCR detection of transgenic Arabidopsis lines (M: DL2000 DNA marker; WT: wild-type Arabidopsis; OE#1-OE#3: Arabidopsis transformed with pBI121-SiSPX9). B: qPCR analysis of transgenic Arabidopsis lines. OE#1-OE#3 indicates overexpression-positive lines. Significant differences were determined by Duncan’s new multiple range test (P<0.05). Different lowercase letters indicate statistically significant differences. Error bars indicate the standard deviation (SD) of three independent biological replicates. Relative expression of the target gene is normalized to Actin. The same below
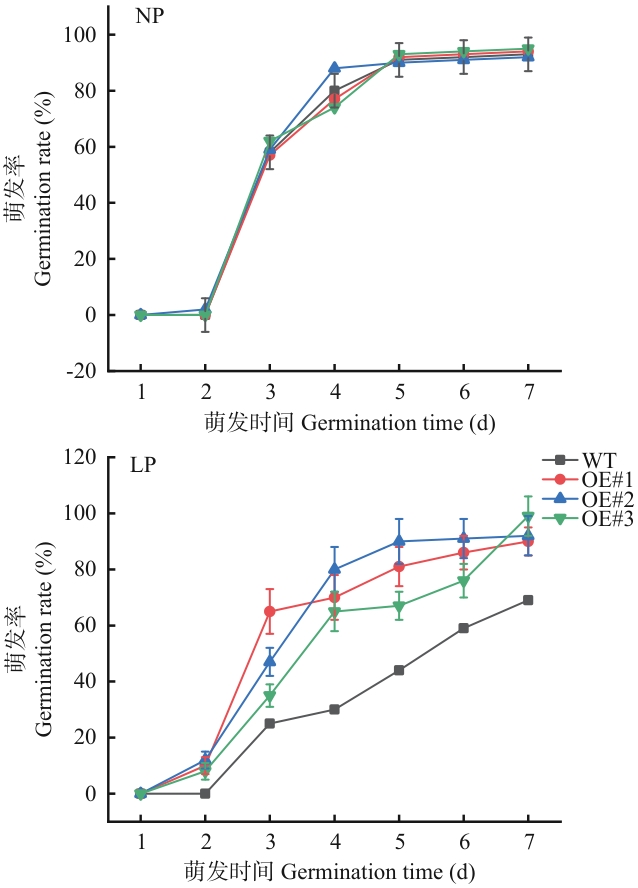
图7 SiSPX9转基因株系萌发率对每次测量使用3个生物学重复进行数据量化,每个重复50粒种子;每个数据点表示3次独立试验的平均值(±标准差)
Fig. 7 Germination rates of SiSPX9 transgenic linesThree biological replicates were used for data quantification for each measurement, with 50 seeds per replicate. Each data point indicates the mean of three independent experiments (± standard deviation)
| [1] | 吕建珍, 任莹, 王宏勇, 等. 264份谷子主要育成品种(系)表型多样性综合评价 [J]. 作物杂志, 2022(4): 22-31. |
| Lü JZ, Ren Y, Wang HY, et al. Comprehensive phenotype evaluation of 264 major foxtail millet bred varieties (lines) [J]. Crops, 2022(4): 22-31. | |
| [2] | 李坤杰. 谷子LBD基因家族分析与SiLBD21功能鉴定 [D]. 杨凌: 西北农林科技大学, 2023. |
| Li KJ. Family analysis of LBD gene in millet and functional identification of SiLBD21 [D]. Yangling: Northwest A & F University, 2023. | |
| [3] | 相吉山, 徐峰, 索良喜, 等. 东北地区谷子地方品种和育成品种表型比较分析 [J]. 植物遗传资源学报, 2018, 19(4): 642-656. |
| Xiang JS, Xu F, Suo LX, et al. Comparison on the phenotypic traits between landraces and cultivars of foxtail millet [Setaria italica (L.) P. beauv.] in Northeast China [J]. J Plant Genet Resour, 2018, 19(4): 642-656. | |
| [4] | 许仙菊, 张永春.植物耐低磷胁迫的根系适应性机制研究进展 [J]. 江苏农业学报, 2018, 34(6): 1425-1429. |
| Xu XJ, & Zhang YC. Research advances in root adaptive mechanisms of plant tolerance to low phosphorus stress [J]. Jiangsu J Agric Sci, 2018, 34(6): 1425-1429. | |
| [5] | 黄沆, 付崇允, 周德贵, 等.植物磷吸收的分子机理研究进展 [J]. 分子植物育种, 2008, (1): 117-122. |
| Huang H, Fu CY, Zhou DG, et al. Research advances in molecular mechanisms of plant phosphorus uptake [J]. Mol Plant Breed, 2008 6(1), 117-122. | |
| [6] | Xu WY, Tang WS, Wang CX, et al. SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway [J]. Front Plant Sci, 2020, 11: 785. |
| [7] | 吴梦洁, 洪家都, 李芳燕, 等. 闽楠SPX基因家族的鉴定及其缺磷胁迫下的表达分析 [J]. 农业生物技术学报, 2023, 31(9): 1832-1845. |
| Wu MJ, Hong JD, Li FY, et al. Identification of SPX gene family in Phoebe bournei and its expression analysis under phosphorus deficiency stress [J]. J Agric Biotechnol, 2023, 31(9): 1832-1845. | |
| [8] | Jung JY, Ried MK, Hothorn M, et al. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain [J]. Curr Opin Biotechnol, 2018, 49: 156-162. |
| [9] | 晋敏姗, 曲瑞芳, 李红英, 等. 谷子糖转运蛋白基因SiSTPs的鉴定及其参与谷子抗逆胁迫响应的研究 [J]. 作物学报, 2022, 48(4): 825-839. |
| Jin MS, Qu RF, Li HY, et al. Identification of sugar transporter gene family SiSTPs in foxtail millet and its participation in stress response [J]. Acta Agron Sin, 2022, 48(4): 825-839. | |
| [10] | Puga MI, Mateos I, Charukesi R, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis [J]. Proc Natl Acad Sci USA, 2014, 111(41): 14947-14952. |
| [11] | 李艳, 方宇辉, 王永霞, 等.不同磷胁迫处理转OsPHR2小麦的转录组学分析 [J]. 作物学报, 2024, 50(2): 340-353. |
| Li Y, Fang YH, Wang YX, et al. Transcriptome analysis of OsPHR2-transgenic wheat under different phosphorus stress treatments [J]. Acta Agron. Sin., 2024, 50(2), 340-353. | |
| [12] | Xiao JB, Xie XM, X, Li C, et al.Identification of SPX family genes in the maize genome and their expression under different phosphate regimes [J]. Plant Biotechnol J, 2021, 168211-220. |
| [13] | 韩长栋. 小麦TaSPX家族鉴定及耐低磷相关基因TaPHO1;H2的功能分析 [D]. 郑州: 河南农业大学, 2023. |
| Han CD. Identification of the TaSPX family in wheat and functional analysis of the low-phosphorus tolerance-related gene TaPHO1;H2 [D]. Zhengzhou: Henan Agricultural University, 2023. | |
| [14] | 商文艳. 小麦SPX基因家族鉴定及TaSPX3在磷胁迫调控中的功能分析 [D]. 郑州: 河南农业大学, 2018. |
| Shang WY. Identification of the wheat SPX gene family and functional analysis of TaSPX3 in phosphorus stress regulation [D]. Zhengzhou: Henan Agricultural University, 2018. | |
| [15] | Wang ZY, Ruan WY, Shi J, et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner [J]. Proc Natl Acad Sci USA, 2014, 111(41): 14953-14958. |
| [16] | Bustos R, Castrillo G, Linhares F, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis [J]. PLoS Genet, 2010, 6(9): e1001102. |
| [17] | Plaxton WC, Tran HT. Metabolic adaptations of phosphate-starved plants [J]. Plant Physiol, 2011, 156(3): 1006-1015. |
| [18] | Wang C, Yue WH, Ying YH, et al. Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice [J]. Plant Physiol, 2015, 169(4): 2822-2831. |
| [19] | Chen JJ, Han XJ, Liu LX, et al. Genome-wide detection of SPX family and profiling of CoSPX-MFS3 in regulating low-phosphate stress in tea-oil Camellia [J]. Int J Mol Sci, 2023, 24(14): 11552. |
| [20] | Luo JL, Liu ZH, Yan JW, et al. Genome-wide identification of SPX family genes and functional characterization of PeSPX6 and PeSPX-MFS2 in response to low phosphorus in Phyllostachys edulis [J]. Plants, 2023, 12(7): 1496. |
| [21] | Duan K, Yi KK, Dang L, et al. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation [J]. Plant J, 2008, 54(6): 965-975. |
| [22] | Pipercevic J, Kohl B, Gerasimaite R, et al. Inositol pyrophosphates activate the vacuolar transport chaperone complex in yeast by disrupting a homotypic SPX domain interaction [J]. Nat Commun, 2023, 14(1): 2645. |
| [23] | Thapa N, Chen M, Horn HT, et al. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4 [J]. Nat Cell Biol, 2020, 22(11): 1357-1370. |
| [24] | 孔晶晶. 玉米ZmLRR1基因调控根系发育的功能和作用机制研究 [D]. 合肥: 安徽农业大学, 2019. |
| Kong JJ. Function and mechanism analysis of ZmLRR1 gene in control of root development in maize [D]. Hefei: Anhui Agricultural University, 2019. | |
| [25] | 李敏. 木薯扩展蛋白家族基因鉴定和功能初步分析 [D]. 海口: 海南大学, 2020. |
| Li M. Genome-wide identification and preliminary functional analysis of expansin gene family in Cassava (Manihot esculenta Crantz) [D]. Haikou: Hainan University, 2020. | |
| [26] | Péret B, Clément M, Nussaume L, et al. Root developmental adaptation to phosphate starvation: better safe than sorry [J]. Trends Plant Sci, 2011, 16(8): 442-450. |
| [27] | 吴敏, 陈瑞, 李贵, 等. 南酸枣苗期耐低磷家系筛选及磷效率评价 [J]. 中南林业科技大学学报, 2025, 45(3): 107-117. |
| Wu M, Chen R, Li G, et al. Screening of low phosphorus tolerant families and phosphorus efficiency of Choerospondias axillaris at seedling stage [J]. J Cent South Univ For Technol, 2025, 45(3): 107-117. | |
| [28] | Dong JS, Piñeros MA, Li XX, et al. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots [J]. Mol Plant, 2017, 10(2): 244-259. |
| [29] | 戴森焕. PHO1介导磷酸盐转运在植物开花调控中的作用机理 [D]. 杭州: 杭州师范大学, 2024. |
| Dai SH. Mechanism of PHO1-mediated phosphate transport in the regulation of plant flowering [D]. Hangzhou: Hangzhou Normal University, 2024. | |
| [30] | 宗伊琳, 彭枫, 袁满, 等. 植物PHT基因家族功能与磷调控网络研究进展 [J]. 上海师范大学学报: 自然科学版, 2024, 53(6): 751-762. |
| Zong YL, Peng F, Yuan M, et al. Research progress on the functions of plant PHT gene family and phosphorus regulatory network [J]. J Shanghai Norm Univ Nat Sci, 2024, 53(6): 751-762. | |
| [31] | Rodríguez MC, Wawrzyńska A, Sirko A. Intronic T-DNA insertion in Arabidopsis NBR1 conditionally affects wild-type transcript level [J]. Plant Signal Behav, 2014, 9(12): e975659. |
| [1] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [2] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [3] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [4] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [5] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [6] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [7] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [8] | 张永艳, 郭思健, 李晶, 郝思怡, 李瑞得, 刘嘉鹏, 程春振. 蓝莓花青素相关VcGSTF19基因的克隆及功能研究[J]. 生物技术通报, 2025, 41(9): 139-146. |
| [9] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [10] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [11] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [12] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [13] | 李开杰, 吴瑶, 李丹丹. 红花CtbHLH128基因克隆及调控干旱胁迫应答功能研究[J]. 生物技术通报, 2025, 41(8): 234-241. |
| [14] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [15] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||