








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 214-225.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0048
收稿日期:2025-01-12
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
刘硕谦,男,博士,教授,研究方向 :茶树分子生物学、生物信息学、品种改良、茶叶品质形成机理;E-mail: shuoqianliu@hunau.edu.cn作者简介:龚钰涵,女,硕士,研究方向 :茶树栽培育种及分子生物学;E-mail: 3166693974@qq.com
基金资助:
GONG Yu-han( ), CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian(
), CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian( )
)
Received:2025-01-12
Published:2025-07-26
Online:2025-07-22
摘要:
目的 端粒结合蛋白(telomere repeat binding, TRB)是一类端粒双链DNA结合蛋白,对植物生长发育起着重要作用。通过鉴定茶树[Camellia sinensis(L.)O. Kuntze]TRB基因家族,克隆关键基因CsTRB1并解析其分子特性,研究CsTRBs在非生物胁迫下的表达模式,为揭示茶树TRB的功能提供分子基础。 方法 对茶树CsTRB基因家族进行全基因组鉴定,通过生物信息学分析保守基序、基因结构、染色体定位、基因共线性以及顺式作用元件,预测蛋白质理化性质及结构。以‘碧香早’为材料,克隆CsTRB1基因CDS序列。结合转录组数据和RT-qPCR对CsTRB基因家族成员在不同组织部位、低温胁迫和激素处理下的表达模式进行分析。 结果 鉴定出7个CsTRBs基因,成功克隆获得CsTRB1基因,其编码区全长885 bp,编码295个氨基酸,包含典型的TRB结构域。家族成员分布在6条染色体上。根据系统发育分析,将CsTRBs分为2个亚家族。共线性分析表明,CsTRB基因在茶树基因组内发生了基因复制事件,茶树TRB家族与拟南芥(Arabidopsis thaliana)的TRB家族之间有7对直系同源基因。RT-qPCR结果表明,在低温胁迫下,CsTRB1、CsTRB2、CsTRB6、CsTRB7显著下降,CsTRB3、CsTRB4、CsTRB5显著上升。除CsTRB5外,茶树CsTRB家族成员皆在ABA处理下高度表达。CsTRB1、CsTRB4、CsTRB5的表达量受到IAA处理的诱导而显著上调。 结论 CsTRBs基因的表达可能受光、低温、激素等顺式作用元件影响,在茶树的生长发育和非生物胁迫响应中发挥重要作用。CsTRB1的成功克隆及其分子特性解析,为后续蛋白互作和功能验证研究提供了关键材料。
龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225.
GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant[J]. Biotechnology Bulletin, 2025, 41(7): 214-225.
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CsTRB1 | TTTGAAGGAGCCTCGTGGG | GTTGCTGCCAACAGCCTTTC |
| CsTRB2 | TGTTGATCTCAAGGACAAATGGAG | ATCCTGTTTAGGGGCAGGCT |
| CsTRB3 | GGCACCTGCCAACTTCAAAC | CCCTCTGCCTTCCCTCTAGT |
| CsTRB4 | TGGTTGTGGCAAAAACACCG | TGCCTCCTTAACGGCTTCAG |
| CsTRB5 | AAAAGCAGAAGTGGACGGCT | CCATGTTCCGCCATTTGTCC |
| CsTRB6 | CTGAATTTGCTCTTTCTCTCACCC | ATTTTGACCATTGGTGAGTGGG |
| CsTRB7 | ATCCGGAATTTAGTGGCGTCT | CTCCATTTGTCCTTGAGATCAACA |
| CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| PCR-CsTRB1 | ATGGTGGTCGCCTCCGGA | CTAAATCCAAATGCTACTGAAGGCT |
表1 引物信息
Table 1 Primer information
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CsTRB1 | TTTGAAGGAGCCTCGTGGG | GTTGCTGCCAACAGCCTTTC |
| CsTRB2 | TGTTGATCTCAAGGACAAATGGAG | ATCCTGTTTAGGGGCAGGCT |
| CsTRB3 | GGCACCTGCCAACTTCAAAC | CCCTCTGCCTTCCCTCTAGT |
| CsTRB4 | TGGTTGTGGCAAAAACACCG | TGCCTCCTTAACGGCTTCAG |
| CsTRB5 | AAAAGCAGAAGTGGACGGCT | CCATGTTCCGCCATTTGTCC |
| CsTRB6 | CTGAATTTGCTCTTTCTCTCACCC | ATTTTGACCATTGGTGAGTGGG |
| CsTRB7 | ATCCGGAATTTAGTGGCGTCT | CTCCATTTGTCCTTGAGATCAACA |
| CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| PCR-CsTRB1 | ATGGTGGTCGCCTCCGGA | CTAAATCCAAATGCTACTGAAGGCT |
基因 ID Gene ID | 基因名 Gene name | 氨基酸数Number of amino acids | 蛋白质分子量 Molecular weight (Da) | 等电点Theoretical pI | 不稳定系数 Instability index | 脂肪指数Aliphatic index | 亲水性 Grand of hydropathicity | 亚细胞定位 Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|
| CSS0029061 | CsTRB1 | 294 | 32 011.53 | 8.56 | 34.83 | 82.52 | -0.497 | 细胞核 |
| CSS0034494 | CsTRB2 | 302 | 33 233.12 | 9.52 | 46.42 | 82.28 | -0.530 | 细胞核 |
| CSS0012155 | CsTRB3 | 300 | 32 813.69 | 9.56 | 49.24 | 81.57 | -0.470 | 细胞核 |
| CSS0030254 | CsTRB4 | 280 | 30 857.84 | 9.16 | 40.08 | 69.79 | -0.688 | 细胞核 |
| CSS0035646 | CsTRB5 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0046266 | CsTRB6 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0022401 | CsTRB7 | 302 | 33 234.06 | 9.52 | 46.42 | 80.99 | -0.557 | 细胞核 |
表2 茶树CsTRBs蛋白物理化学性质
Table 2 Physicochemical properties of CsTRBs proteins in Camellia sinensis
基因 ID Gene ID | 基因名 Gene name | 氨基酸数Number of amino acids | 蛋白质分子量 Molecular weight (Da) | 等电点Theoretical pI | 不稳定系数 Instability index | 脂肪指数Aliphatic index | 亲水性 Grand of hydropathicity | 亚细胞定位 Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|
| CSS0029061 | CsTRB1 | 294 | 32 011.53 | 8.56 | 34.83 | 82.52 | -0.497 | 细胞核 |
| CSS0034494 | CsTRB2 | 302 | 33 233.12 | 9.52 | 46.42 | 82.28 | -0.530 | 细胞核 |
| CSS0012155 | CsTRB3 | 300 | 32 813.69 | 9.56 | 49.24 | 81.57 | -0.470 | 细胞核 |
| CSS0030254 | CsTRB4 | 280 | 30 857.84 | 9.16 | 40.08 | 69.79 | -0.688 | 细胞核 |
| CSS0035646 | CsTRB5 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0046266 | CsTRB6 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0022401 | CsTRB7 | 302 | 33 234.06 | 9.52 | 46.42 | 80.99 | -0.557 | 细胞核 |

图1 茶树CsTRB与其他TRB家族蛋白氨基酸序列比对图中红线标记为MYB结构域,黄线标记为组蛋白H1/5结构域,蓝线标记为卷曲螺旋结构域;Cs:茶树;At:拟南芥;Os:水稻;Zm:玉米
Fig. 1 Amino acid sequence alignment of CsTRB from tea plant with other TRB family proteinsThe MYB domain is marked with a red line, the histone H1/5 domain is marked with a yellow line, and the coil domain is marked with a blue line. Cs: Camellia sinensis. At: Arabidopsis thaliana. Os: Oryza sativa. Zm: Zea mays
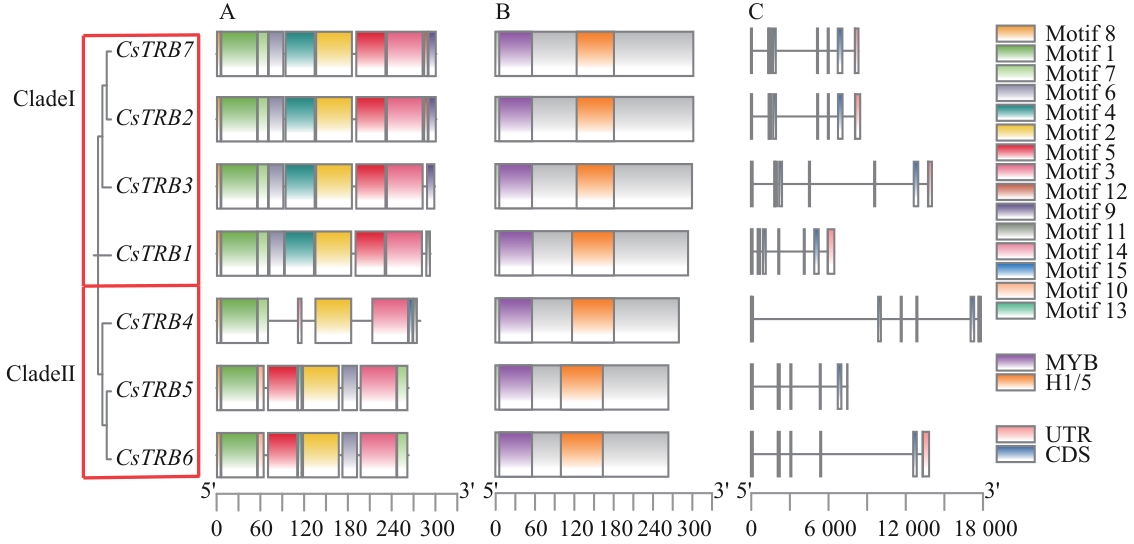
图4 茶树TRB家族基因基序、CDD及结构分析A:基于系统发育关系和结构域鉴定,将CsTRBs划分为2个支系;B:CsTRB蛋白中的保守基序组成,不同颜色的盒子表示不同的结构域;C:CsTRBs基因结构,蓝色矩形表示CDS或外显子,灰色线表示内含子
Fig. 4 Motif, CDD and structural analysis of TRB family genes in C. sinensisA: Identification based on phylogenetic relationships and domain, CsTRBs were classified into two subgroups. B: Composition of conserved motifs within CsTRB proteins. Boxes of different colors indicate distinct domains. C: Gene structures of CsTRBs. Blue rectangles denote CDS or exons, while gray lines indicate introns
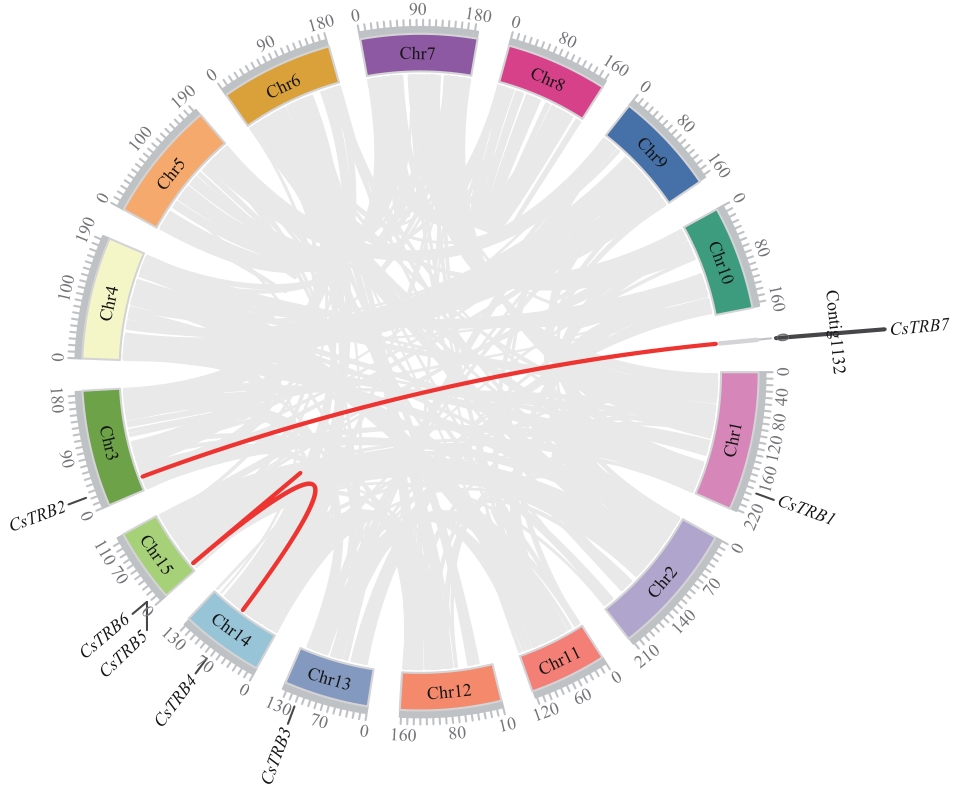
图5 CsTRB基因的染色体定位和染色体间关联背景中的灰线显示了茶树基因组中所有的synsynblock,红线显示了synsynbnlor基因对
Fig. 5 Chromosomal localization and inter-chromosomal associations of CsTRB genesIn the background, gray lines indicate all the synsynblocks in the tea plant genome, while red lines indicate the synsynbnlor gene pairs

图6 茶树和拟南芥中TRB基因家族的共线性分析背景中的灰线表示茶树和拟南芥的共线性片段,蓝线表示共线性的TRB基因对
Fig. 6 Synteny analysis of TRB gene family between C. sinensis and A. thalianThe gray lines in the background depict syntenic blocks between the tea plant and Arabidopsis, while blue lines indicate syntenic TRB gene pairs

图9 CsTRB基因转录组表达谱A、C、D、E:分别表示CsTRB基因在不同组织、200 mmol/L NaCl、25% PEG和100 mmol/L MeJA胁迫下的表达分析,数据使用log2(FPKM+1)转换进行转换,红色方块表示基因被上调,蓝色方块表示基因被下调;B:茶树CsTRB基因在低温驯化下的表达分析(CK:25 ℃;Cold1-6 h:10 ℃,6 h;Cold1-7 d:昼/夜10 ℃/4 ℃,持续7 d;Cold2-7 d:昼/夜4 ℃/0 ℃ 7 d;ColdDA-7 d:在25 ℃,恢复7 d)
Fig. 9 Transcriptome expression profile of CsTRB genesA, C, D, and E: Indicate the expression analysis of CsTRB gene in different tissues, 200 mmol/L NaCl, 25% PEG and 100 mmol/L MeJA stress, the data were converted by log2 (FPKM+1), the red square indicates that the gene was up-regulated, and the blue square indicates that the gene was down-regulated. B: Expression analysis of CsTRB gene in tea plant under low temperature acclimation (CK: Non-acclimated at 25 ℃; Cold1-6 h: fully acclimated at 10 ℃ for 6 h; Cold1-7 d: 10 ℃/4 ℃ at day/night for 7 d; Cold2-7 d: 4 ℃/0 ℃ at day/night for 7 d; ColdDA-7 d: recovering under 25 ℃ for 7 d)

图10 CsTRB基因在不同非生物胁迫下的表达谱A:CsTRBs基因在低温胁迫下的表达谱;B:CsTRBs基因在100 μmol/L ABA处理下的表达谱;C:CsTRBs基因在100 μmol/L IAA处理下的表达谱。误差线代表3次独立生物学重复标准偏差;不同小写字母表示同一种处理不同时间点差异显著(P<0.05)
Fig. 10 Expression profile of CsTRB gene under different abiotic stressesA: Expression profile of CsTRBs gene under low temperature stress. B: Expression profile of CsTRBs gene under 100 μmol/L ABA treatment. C: Expression profile of the CsTRBs gene under 100 μmol/L IAA treatment. The error bars indicate the standard deviation of 3 independent biological replicates. Different lowercase letters indicate significant differences at different time points of the same treatment (P<0.05)
| [1] | Riha K, Shippen DE. Telomere structure, function and maintenance in Arabidopsis [J]. Chromosome Res, 2003, 11(3): 263-275. |
| [2] | Bilaud T, Koering CE, Binet-Brasselet E, et al. The telobox, a MYB-related telomeric DNA binding motif found in proteins from yeast, plants and human [J]. Nucleic Acids Res, 1996, 24(7): 1294-1303. |
| [3] | Peška V, Schrumpfová PP, Fajkus J. Using the telobox to search for plant telomere binding proteins [J]. Curr Protein Pept Sci, 2011, 12(2): 75-83. |
| [4] | Kuchar M, Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants [J]. FEBS Lett, 2004, 578(3): 311-315. |
| [5] | Schrumpfová P, Kuchar M, Miková G, et al. Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence [J]. Genome, 2004, 47(2): 316-324. |
| [6] | Marian CO, Bordoli SJ, Goltz M, et al. The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro [J]. Plant Physiol, 2003, 133(3): 1336-1350. |
| [7] | Amiard S, Feit L, Simon L, et al. Distinct clades of telomere repeat binding transcriptional regulators interplay to regulate plant development [J/OL]. bioRxiv, 2023. DOI:10.1101/2023.08.16.553498 . |
| [8] | He Q, Chen L, Xu Y, et al. Identification of centromeric and telomeric DNA-binding proteins in rice [J]. Proteomics, 2013, 13(5): 826-832. |
| [9] | Yang SW, Kim DH, Lee JJ, et al. Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells [J]. J Biol Chem, 2003, 278(24): 21395-21407. |
| [10] | 张冬杰, 燕丽萍, 吴其超, 等. 绒毛白蜡smh1基因编码区的克隆及序列分析 [J]. 中国农学通报, 2017, 33(15): 49-55. |
| Zhang DJ, Yan LP, Wu QC, et al. Cloning and sequence analysis of Fraxinus velutina smh1 gene coding region [J]. Chin Agric Sci Bull, 2017, 33(15): 49-55. | |
| [11] | Mozgová I, Schrumpfová PP, Hofr C, et al. Functional characterization of domains in AtTRB1, a putative telomere-binding protein in Arabidopsis thaliana [J]. Phytochemistry, 2008, 69(9): 1814-1819. |
| [12] | Hofr C, Sultesová P, Zimmermann M, et al. Single-myb-histone proteins from Arabidopsis thaliana: a quantitative study of telomere-binding specificity and kinetics [J]. Biochem J, 2009, 419(1): 221-230. |
| [13] | Schrumpfová PP, Vychodilová I, Dvořáčková M, et al. Telomere repeat binding proteins are functional components of Arabidopsis telomeres and interact with telomerase [J]. Plant J, 2014, 77(5): 770-781. |
| [14] | Chen YH, Yang XY, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family [J]. Plant Mol Biol, 2006, 60(1): 107-124. |
| [15] | An JP, Xu RR, Liu X, et al. Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex [J]. Plant J, 2021, 106(5): 1414-1430. |
| [16] | Zhou Y, Wang YJ, Krause K, et al. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis [J]. Nat Genet, 2018, 50(5): 638-644. |
| [17] | Hong JP, Byun MY, Koo DH, et al. Suppression of RICE TELOMERE BINDING PROTEIN 1 results in severe and gradual developmental defects accompanied by genome instability in rice [J]. Plant Cell, 2007, 19(6): 1770-1781. |
| [18] | Procházková Schrumpfová P, Schořová Š, Fajkus J. Telomere- and telomerase-associated proteins and their functions in the plant cell [J]. Front Plant Sci, 2016, 7: 851. |
| [19] | 莫晓丽, 黄亚辉. 茶树主要逆境胁迫反应及其适应逆境的生理机制 [J]. 茶叶学报, 2021, 62(4): 185-190. |
| Mo XL, Huang YH. Responses and resistance mechanisms of tea plants to stresses—a review [J]. Acta Tea Sin, 2021, 62(4): 185-190. | |
| [20] | 王鹏杰, 杨江帆, 张兴坦, 等. 茶树基因组与测序技术的研究进展 [J]. 茶叶科学, 2021, 41(6): 743-752. |
| Wang PJ, Yang JF, Zhang XT, et al. Research advance of tea plant genome and sequencing technologies [J]. J Tea Sci, 2021, 41(6): 743-752. | |
| [21] | El-Gebali S, Mistry J, Bateman A, et al. The pfam protein families database in 2019 [J]. Nucleic Acids Res, 2019, 47(D1): D427-D432. |
| [22] | Potter SC, Luciani A, Eddy SR, et al. HMMER web server: 2018 update [J]. Nucleic Acids Res, 2018, 46(w1): W200-W204. |
| [23] | Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0 [J]. Bioinformatics, 2007, 23(21): 2947-2948. |
| [24] | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| [25] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [26] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [27] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [28] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics [J]. Genome Res, 2009, 19(9): 1639-1645. |
| [29] | Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice [J]. BMC Plant Biol, 2010, 10: 282. |
| [30] | Pontius J, Richelle J, Wodak SJ. Deviations from standard atomic volumes as a quality measure for protein crystal structures [J]. J Mol Biol, 1996, 264(1): 121-136. |
| [31] | Delano WL. Pymol: An open-source molecular graphics tool [J]. CCP4 Newsletter on Protein Crystallography, 2002, 40(1): 82-92. |
| [32] | Swift ML. GraphPad prism, data analysis, and scientific graphing [J]. J Chem Inf Comput Sci, 1997, 37(2): 411-412. |
| [33] | Wang L, Ding XL, Gao YQ, et al. Genome-wide identification and characterization of GRAS genes in soybean (Glycine max) [J]. BMC Plant Biol, 2020, 20(1): 415. |
| [34] | Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure [J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192. |
| [35] | Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain [J]. Proc Natl Acad Sci USA, 2003, 100(12): 7158-7162. |
| [36] | Rose AB. Introns as gene regulators: a brick on the accelerator [J]. Front Genet, 2019, 9: 672. |
| [37] | Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor [J]. Trends Genet, 2008, 24(8): 375-378. |
| [38] | Chen HY, Hsieh EJ, Cheng MC, et al. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element [J]. New Phytol, 2016, 211(2): 599-613. |
| [39] | Zhou Y, Hartwig B, James GV, et al. Complementary activities of telomere repeat binding proteins and polycomb group complexes in transcriptional regulation of target genes [J]. Plant Cell, 2016, 28(1): 87-101. |
| [40] | Kusová A, Steinbachová L, Přerovská T, et al. Completing the TRB family: newly characterized members show ancient evolutionary origins and distinct localization, yet similar interactions [J]. Plant Mol Biol, 2023, 112(1/2): 61-83. |
| [1] | 张学琼, 潘素君, 李魏, 戴良英. 植物磷酸盐转运蛋白在胁迫响应中的研究进展[J]. 生物技术通报, 2025, 41(7): 28-36. |
| [2] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [3] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [4] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [5] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [6] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [7] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [8] | 吴浩, 董伟峰, 贺子天, 李艳肖, 谢辉, 孙明哲, 沈阳, 孙晓丽. 水稻BXL基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(6): 87-98. |
| [9] | 瞿美玲, 周思敏, 张惊宇, 何佳蔚, 朱佳源, 刘笑蓉, 童巧珍, 周日宝, 刘湘丹. 灰毡毛忍冬bHLH转录基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(6): 256-268. |
| [10] | 黄丹, 彭兵阳, 张盼盼, 焦悦, 吕佳斌. 油茶HD-Zip基因家族鉴定及其在非生物胁迫下的表达分析[J]. 生物技术通报, 2025, 41(6): 191-207. |
| [11] | 刘鑫, 王嘉雯, 李进伟, 牟策, 杨盼盼, 明军, 徐雷锋. 兰州百合三个LdBBXs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(5): 186-196. |
| [12] | 彭绍智, 王登科, 张祥, 戴雄泽, 徐昊, 邹学校. 辣椒CaFD1基因克隆、表达特征及功能验证[J]. 生物技术通报, 2025, 41(5): 153-164. |
| [13] | 叶柳健, 蒙健宗, 覃福方, 何双, 朱绮霞, 王小虎, 韦圣博, 周礼芹. 古茶树林菌株D2的鉴定、酶学特性及基因组学分析[J]. 生物技术通报, 2025, 41(5): 267-279. |
| [14] | 赵婧, 郭茜, 李睿琦, 雷滢炀, 岳爱琴, 赵晋忠, 殷丛丛, 杜维俊, 牛景萍. 大豆GmGST基因簇基因序列分析及诱导表达分析[J]. 生物技术通报, 2025, 41(5): 129-140. |
| [15] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||