








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 225-239.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0416
张晓丹1, 尹铮1( ), 刘清晨1, 李雪梅2, 刘晓华1(
), 刘清晨1, 李雪梅2, 刘晓华1( ), 梁美霞1(
), 梁美霞1( )
)
收稿日期:2025-04-20
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
刘晓华,女,博士,讲师,研究方向 :园艺植物观赏性状与抗逆机理;E-mail: 3305@ldu.edu.cn作者简介:张晓丹,女,硕士,研究方向 :园艺植物耐逆机理;E-mail: 1491091164@qq.com
基金资助:
ZHANG Xiao-dan1, YIN Zheng1( ), LIU Qing-chen1, LI Xue-mei2, LIU Xiao-hua1(
), LIU Qing-chen1, LI Xue-mei2, LIU Xiao-hua1( ), LIANG Mei-xia1(
), LIANG Mei-xia1( )
)
Received:2025-04-20
Published:2025-12-26
Online:2026-01-06
摘要:
目的 冷冻胁迫是限制桃树生长、果实品质和产量的主要环境因素之一。然而,关于桃树应对低温响应的分子机制目前仍知之甚少,探究低温胁迫下桃树氨基酸、碳水化合物及脂质代谢的动态重编程规律及其与抗寒性的分子关联,为抗寒分子育种及栽培技术优化提供理论支撑。 方法 以山桃(Prunus davidiana)为材料,采用梯度冷冻低温处理(5、-5、-15、-25 ℃),测定生理指标并结合转录组学与代谢组学的比较分析,通过整合生理生化测定、代谢组学和转录组学技术,系统解析其多组学调控网络。 结果 冷冻胁迫触发山桃脯氨酸与可溶性糖的积累,伴随丙二醛(MDA)含量及电解质渗漏率(EL)升高,表明细胞膜系统受损。代谢组学分析显示,低温下糖类及其衍生物显著富集,部分氨基酸减少,而三羧酸循环(TCA)相关有机酸(如2-氧代戊二酸、瓜氨酸)含量增加。黄酮类生物合成、精氨酸合成及氮代谢通路在胁迫中显著激活。转录组分析证实,葡萄糖苷酸生物合成相关基因在低温下表达显著上调,与代谢物变化协同响应冷冻胁迫,揭示山桃通过代谢‒基因网络调控增强抗寒能力的分子机制。 结论 揭示山桃应对冷冻胁迫时激活淀粉与蔗糖代谢、苯丙烷及黄酮类合成途径,筛选出21种关键代谢物和15个核心基因,阐明了抗冻代谢与基因调控协同机制。
张晓丹, 尹铮, 刘清晨, 李雪梅, 刘晓华, 梁美霞. 基于代谢组和转录组联合解析山桃响应冻害机制[J]. 生物技术通报, 2025, 41(12): 225-239.
ZHANG Xiao-dan, YIN Zheng, LIU Qing-chen, LI Xue-mei, LIU Xiao-hua, LIANG Mei-xia. Integrated Metabolomic and Transcriptomic Analysis Reveals the Mechanism of Prunus davidiana Response to Freezing Stress[J]. Biotechnology Bulletin, 2025, 41(12): 225-239.

图1 冷冻温度胁迫下山桃表型(A)及生理指标分析(B、C)数据为3次独立试验的平均值±标准差。不同字母表示经单因素方差分析(ANOVA)及Duncan多重比较检验确定的组间差异显著性(P<0.05)。CK:5 ℃;T5:-5 ℃;T15:-15 ℃;T25:-25 ℃。下同
Fig. 1 Analysis of phenotypic (A) and physiological indices (B, C) of P. davidiana under freezing temperature stressData are means ± standard deviation of three independent experiments. Different letters indicate the significance of differences between groups as determined by one-way analysis of variance (ANOVA) and Duncan’s multiple comparison test (P<0.05). The same below

图2 山桃在不同冷冻胁迫下的差异积累代谢物分析A:不同温度下鉴定代谢物的主成分分析(PCA);B:4种不同温度处理下所有差异代谢物(DEMs)的热图,颜色表示各差异代谢物的相对含量水平,红色表示高含量,绿色表示低含量;C:CK vs T5、CK vs T15和CK vs T25差异代谢物的火山图;D:所有差异代谢物根据变化趋势进行的聚类分析,详细聚类结果见图S2;E:CK vs T5、CK vs T15和CK vs T25差异代谢物的维恩图;F:CK vs T5、CK vs T15和CK vs T25 3组共有109个差异代谢物的热图,详细差异代谢物结果见表S5
Fig. 2 Analysis of differentially accumulated metabolites in P. davidiana under different freezing stressesA: Principal component analysis (PCA) of the identified metabolites at different temperatures. B: Heat maps of all differential metabolites (DEMs) at four different temperature treatments. Colours indicate the relative content level of each differential metabolite, with red indicating high content and green indicating low content. C: Volcano plots of CK vs T5, CK vs T15 and CK vs T25 differential metabolites. D: Cluster analysis of all differential metabolites according to the trend of changes, and the detailed clustering results are shown in Figure S2. E: Venn diagrams of CK vs T5, CK vs T15 and CK vs T25 differential metabolites. F: Heat maps of a total of 109 differential metabolites in the three groups of CK vs T5, CK vs T15 and CK vs T25, and the detailed results of differential metabolites are shown in Table S5

图3 差异表达代谢物(DEMs)的KEGG通路富集分析A-C:CK vs T5、CK vs T15和CK vs T25比较分析的KEGG 通路富集分析图;D-F:CK vs T5、CK vs T15和CK vs T25比较分析前10种差异表达代谢物
Fig. 3 KEGG pathway enrichment analysis of differentially expressed metabolites (DEMs)A-C: KEGG pathway enrichment analysis of the comparisons CK vs T5, CK vs T15, and CK vs T25. D-F: Top 10 differentially expressed metabolites in the comparing CK vs T5, CK vs T15, and CK vs T25
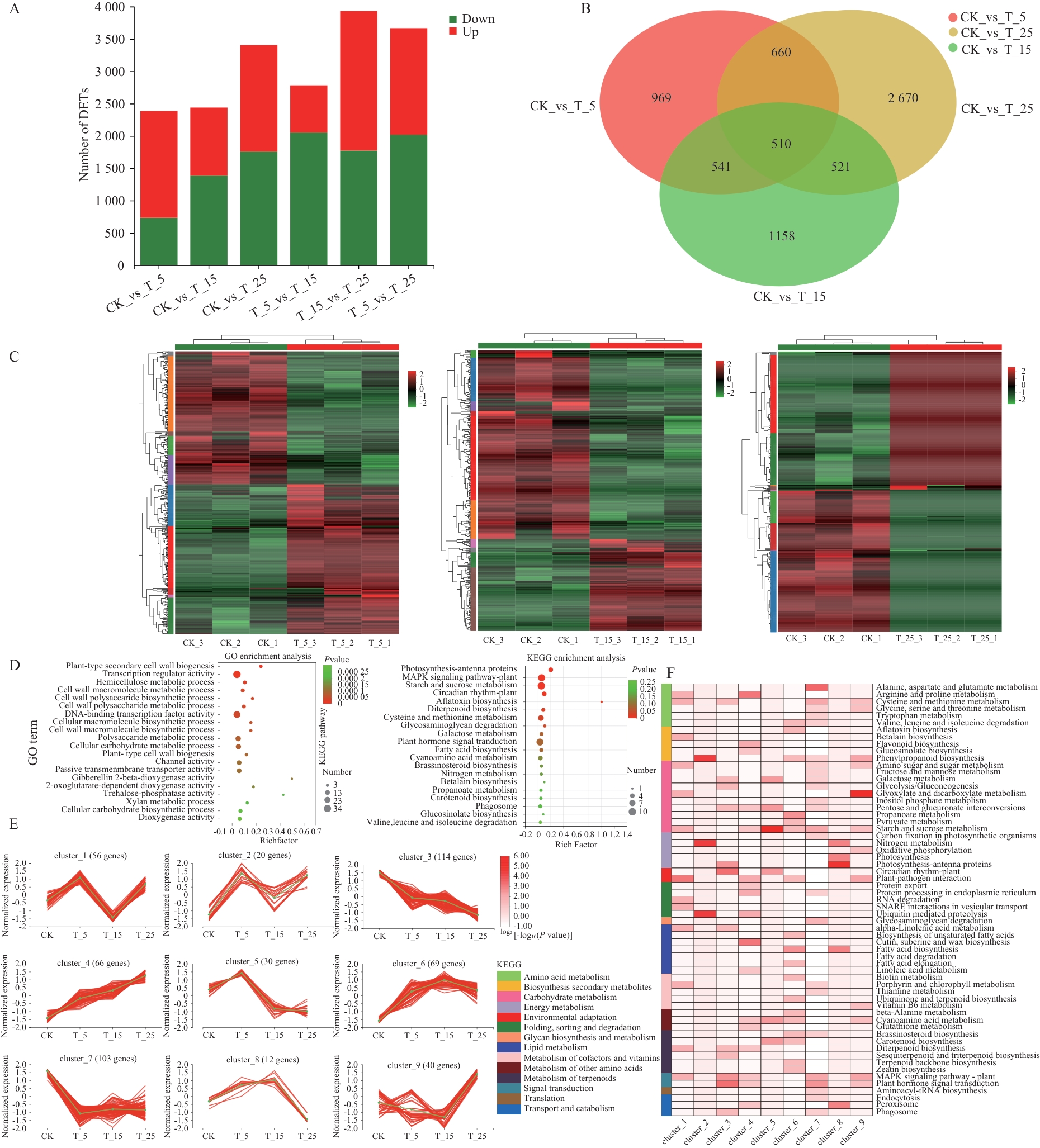
图4 差异表达基因(DEGs)表达模式及共表达基因的GO与KEGG富集分析A:DEGs上调和下调情况;B:不同处理组间DEGs表达Venn图;C:所有比较组中DEGs的共调控关系;D:510个共表达基因的前20位KEGG和GO富集分析结果;E:510个共表达DEGs被划分为9个聚类,每个聚类的基因数量显示在顶部,绿色线条表示各子聚类相对表达水平的平均值,橘色线条表示各子聚类中单个基因的相对表达水平;F:各聚类中显著富集的KEGG通路(P<0.05)
Fig. 4 Expression patterns of differentially expressed genes (DEGs) and GO and KEGG enrichment analyses of co-expressed genesA: Up- and down-regulation of DEGs. B: Venn map of DEGs expression among different treatment groups. C: Co-regulatory relationships of DEGs in all comparison groups. D: Results of KEGG and GO enrichment analyses of top 20 KEGGs and GOs for the 510 co-expressed genes. E: The 510 co-expressed DEGs were classified into 9 clusters. The number of genes in each cluster is shown at the top, the green line indicates the average of the relative expression in each subcluster, and the pink line indicates the relative expressions of individual genes in each subcluster. F: KEGG pathways significantly enriched in each cluster (P<0.05)
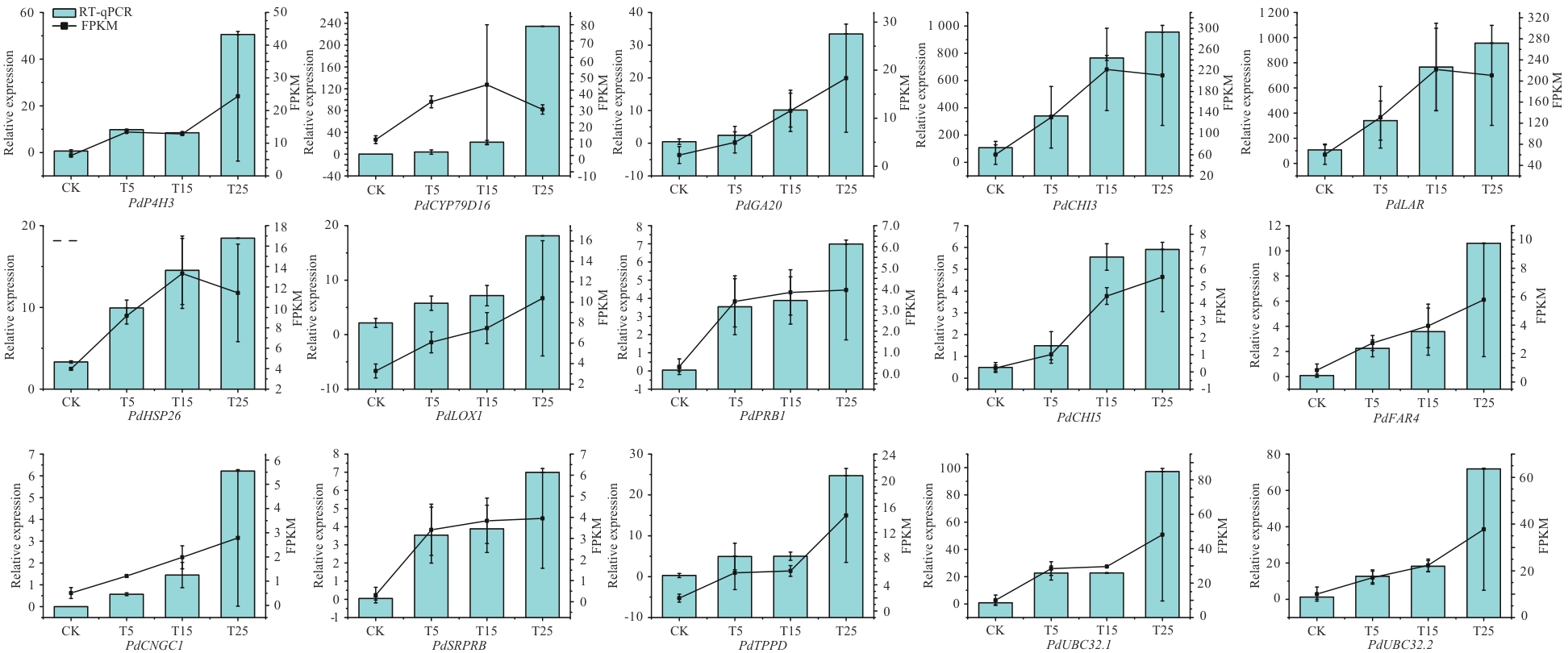
图5 RNA-Seq数据的RT-qPCR验证数据表示为3个独立生物学重复的平均值±标准误
Fig. 5 RT-qPCR validation of RNA-Seq dataData showed the means ± SE of three independent biological replicates
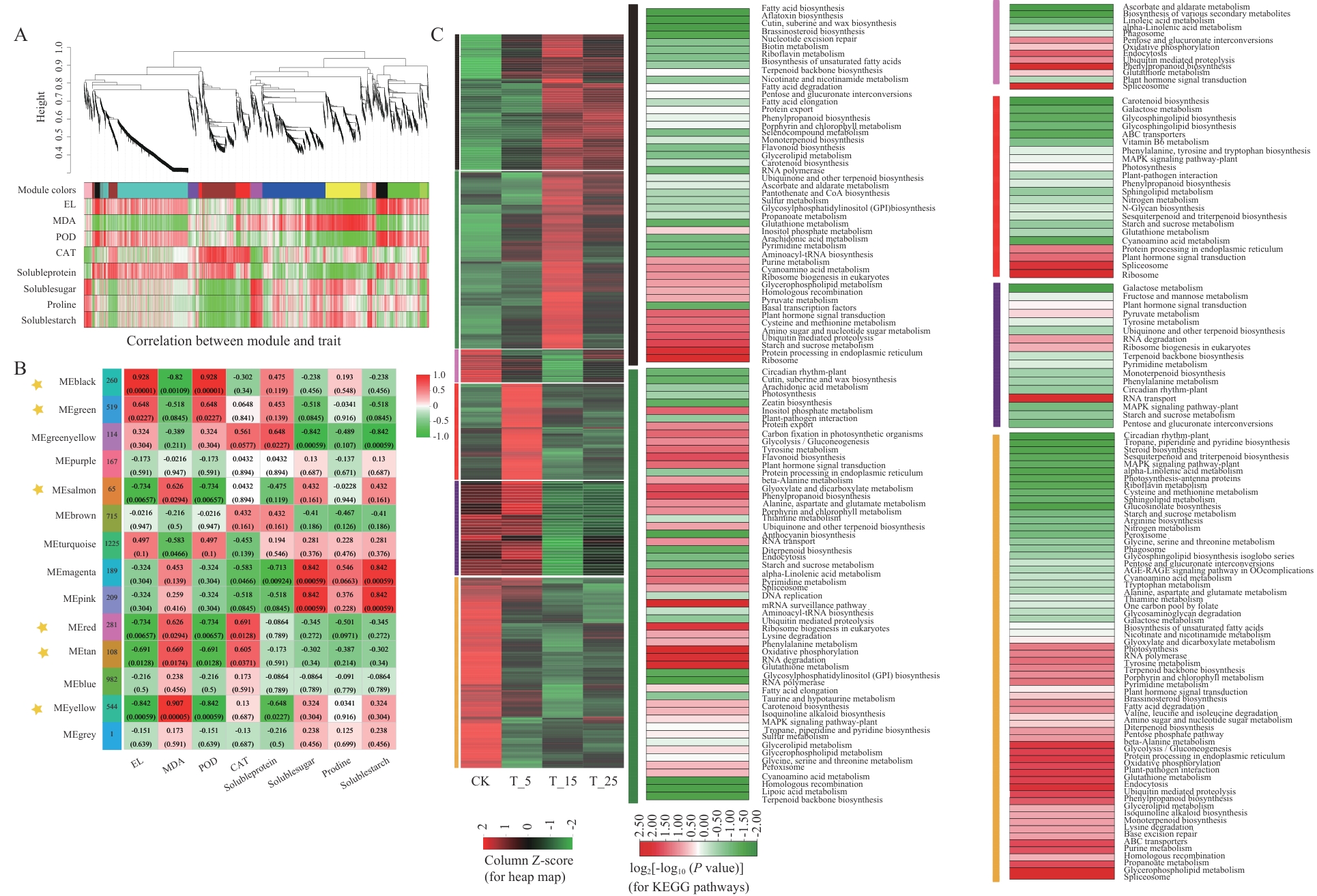
图6 基因WGCNA分析与生理性状的关联性A:基于WGCNA的层次聚类树分析,主分支划分为14个不同颜色模块;B:模块‒性状相关性热图,每行对应1个模块(颜色与A图一致),行列交叉处的颜色反映模块与生理性状的相关性,标有五角星的黑、绿、鲑红、红、棕褐及黄色模块与电解质渗漏率(EL)、丙二醛(MDA)及过氧化物酶活性(POD)显著相关(P<0.05);C:上述6个模块中DEGs的温度响应趋势及KEGG通路富集分析(通过R语言pHYPER函数实现,P<0.05)
Fig. 6 Association of gene WGCNA analysis with physiological traitsA: Hierarchical clustering tree analysis based on WGCNA, with the main branch divided into 14 modules of different colours. B: Heatmap of module-trait correlation, each row corresponds to one module (colours are consistent with Figure A), and the colours at the intersections of the rows and columns reflect the correlation between the modules and the physiological traits, the black, green, salmon red, red, brownish-brown and yellow modules labelled with an asterisk are significantly (P<0.05) correlated with the electrolyte leakage rate (EL), malondialdehyde (MDA) and peroxidase activity (POD) are significantly correlated (P<0.05). C: Temperature response trend and KEGG pathway enrichment analysis of DEGs in the above six modules (achieved by R language pHYPER function, P<0.05)
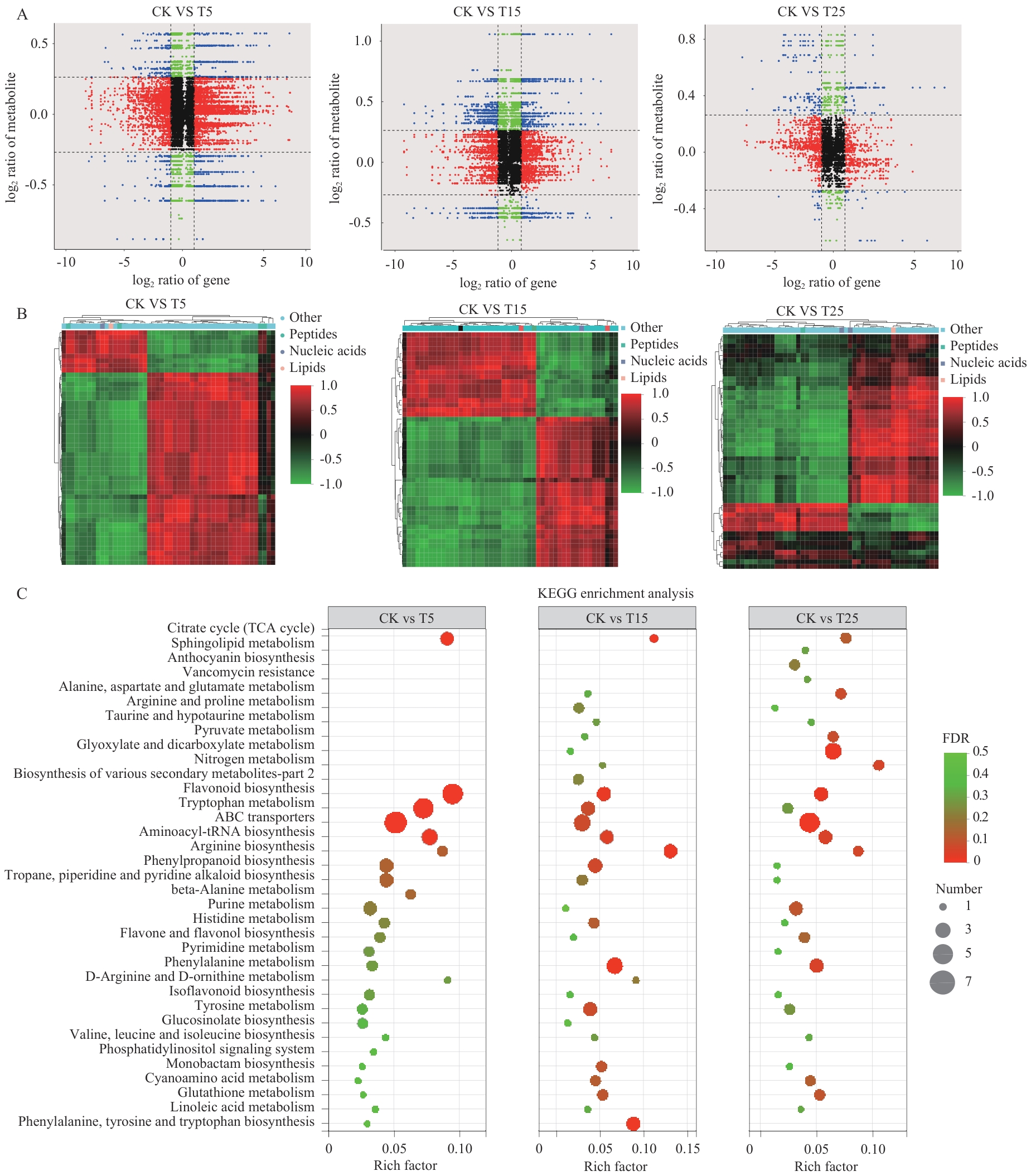
图7 山桃响应不同强度冷冻胁迫的代谢组与转录组关联分析A:九象限图展示代谢物与基因的关联性,红色代表基因和代谢物均上调,绿色代表基因和代谢物均下调,黑色代表表示统计不显著的关联,蓝色表示基因与代谢物表达趋势相反;B:处理组(CK vs T5/T15/T25)与对照组间DAMs的相关系数聚类热图(PCCs>0.8);C:CK vs T5/T15/T25 3组比较中DEMs前20位KEGG富集通路汇总分析
Fig. 7 Metabolome-transcriptome association analysis of P. davidiana in response to different intensities of freezing stressA: Nine-quadrant plot demonstrates metabolite-gene associations, red indicates that both genes and metabolites are upregulated, green indicates that both genes and metabolites are downregulated, black indicates a statistically insignificant association, and blue indicates that the expression trends of genes and metabolites are opposite. B: Clustered heatmap of correlation coefficients of DAMs (PCCs>0.8) between treatment (CK vs T5/T15/T25) and control groups. C: Top 20 DEMs in the comparison of the three groups CK vs T5/T15/T25 KEGG-enriched pathway summary analysis

图8 冷冻胁迫下山桃差异表达代谢物(DEMs)与差异表达基因(DEGs)参与的代谢通路A:淀粉与糖代谢;B:有机酸代谢;C;类黄酮合成。圆形表示受调控代谢物(绿色下调,红色上调),矩形表示受调控基因(绿色下调,红色上调)
Fig. 8 Metabolic pathways involved in differentially expressed metabolites (DEMs) and differentially expressed genes (DEGs) in P. davidiana under freezing stressA: Starch and sugar metabolism. B: Organic acid metabolism. C: Flavonoid synthesis. Circles indicate regulated metabolites (green down-regulation, red up-regulation), rectangles indicate regulated genes (green down-regulation, red up-regulation)
| [1] | Wang K, Yin XR, Zhang B, et al. Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit [J]. Plant Cell Environ, 2017, 40(8): 1531-1551. |
| [2] | Ding YL, Shi YT, Yang SH. Regulatory networks underlying plant responses and adaptation to cold stress [J]. Annu Rev Genet, 2024, 58(1): 43-65. |
| [3] | Yu QH, Zheng QL, Liu C, et al. Phosphorylation-dependent VaMYB4a regulates cold stress in grapevine by inhibiting VaPIF3 and activating VaCBF4 [J]. Plant Physiol, 2025, 197(2): kiaf035. |
| [4] | Liu XY, Bulley SM, Varkonyi-Gasic E, et al. Kiwifruit bZIP transcription factor AcePosF21 elicits ascorbic acid biosynthesis during cold stress [J]. Plant Physiol, 2023, 192(2): 982-999. |
| [5] | Souza AG, Smiderle OJ, Menegatti RD, et al. Patents for the physiological quality in seeds of peach rootstock classified by weight and stored for different periods [J]. Recent Pat Food Nutr Agric, 2019, 10(2): 124-130. |
| [6] | Javed Tareen M, Wang XK, Ali I, et al. Influence of Scion/Rootstock reciprocal effects on post-harvest and metabolomics regulation in stored peaches [J]. Saudi J Biol Sci, 2022, 29(1): 427-435. |
| [7] | Cao K, Peng Z, Zhao X, et al. Chromosome-level genome assemblies of four wild peach species provide insights into genome evolution and genetic basis of stress resistance [J]. BMC Biol, 2022, 20(1): 139. |
| [8] | Guo ML, Pan RJ, Chu ZJ, et al. Low-temperature stress response: a transcriptomic study of the WRKY family in Prunus davidiana [J]. Cryobiology, 2025, 119: 105252. |
| [9] | Niu RX, Zhao XM, Wang CB, et al. Transcriptome profiling of Prunus persica branches reveals candidate genes potentially involved in freezing tolerance [J]. Sci Hortic, 2020, 259: 108775. |
| [10] | Li YH, Tian QH, Wang ZY, et al. Integrated analysis of transcriptomics and metabolomics of peach under cold stress [J]. Front Plant Sci, 2023, 14: 1153902. |
| [11] | Muthuramalingam P, Shin H, Adarshan S, et al. Molecular insights into freezing stress in peach based on multi-omics and biotechnology: an overview [J]. Plants, 2022, 11(6): 812. |
| [12] | Wang WB, Kim YH, Lee HS, et al. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses [J]. Plant Physiol Biochem, 2009, 47(7): 570-577. |
| [13] | Nguyen NH, Van Le B. A simple, economical, and high efficient protocol to produce in vitro miniature rose [J]. Vitro Cell Dev Biol Plant, 2020, 56(3): 362-365. |
| [14] | Wen B, Mei ZL, Zeng CW, et al. metaX: a flexible and comprehensive software for processing metabolomics data [J]. BMC Bioinformatics, 2017, 18(1): 183. |
| [15] | Maruyama K, Urano K, Yoshiwara K, et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts [J]. Plant Physiol, 2014, 164(4): 1759-1771. |
| [16] | MAN Sambe, He XY, Tu QH, et al. A cold-induced myo-inositol transporter-like gene confers tolerance to multiple abiotic stresses in transgenic tobacco plants [J]. Physiol Plant, 2015, 153(3): 355-364. |
| [17] | Negrão S, Schmöckel SM, Tester M. Evaluating physiological responses of plants to salinity stress [J]. Ann Bot, 2017, 119(1): 1-11. |
| [18] | Liu B, Zhao FM, Cao Y, et al. Photoprotection contributes to freezing tolerance as revealed by RNA-seq profiling of Rhododendron leaves during cold acclimation and deacclimation over time [J]. Hortic Res, 2022, 9: uhab025. |
| [19] | Hao XY, Wang B, Wang L, et al. Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves [J]. Sci Hortic, 2018, 240: 354-368. |
| [20] | Lu WJ, Wei GQ, Zhou BW, et al. A comparative analysis of photosynthetic function and reactive oxygen species metabolism responses in two Hibiscus cultivars under saline conditions [J]. Plant Physiol Biochem, 2022, 184: 87-97. |
| [21] | Mellado-Mojica E, Calvo-Gómez O, Jofre-Garfias AE, et al. Fructooligosaccharides as molecular markers of geographic origin, growing region, genetic background and prebiotic potential in strawberries: a TLC, HPAEC-PAD and FTIR study [J]. Food Chem Adv, 2022, 1: 100064. |
| [22] | Yoon J, Cho LH, Tun W, et al. Sucrose signaling in higher plants [J]. Plant Sci, 2021, 302: 110703. |
| [23] | Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants [J]. J Biosci, 2005, 30(5): 761-776. |
| [24] | Fait A, Fromm H, Walter D, et al. Highway or byway: the metabolic role of the GABA shunt in plants [J]. Trends Plant Sci, 2008, 13(1): 14-19. |
| [25] | Liang X, Wang Y, Shen WX, et al. Genomic and metabolomic insights into the selection and differentiation of bioactive compounds in Citrus [J]. Mol Plant, 2024, 17(11): 1753-1772. |
| [26] | Wang YL, Tong W, Li FD, et al. LUX ARRHYTHMO links CBF pathway and jasmonic acid metabolism to regulate cold tolerance of tea plants [J]. Plant Physiol, 2024, 196(2): 961-978. |
| [27] | Amjadi E, Ganjeali A, Shakeri A, et al. Ecophysiological and phytochemical insights in to Silybum marianum: geographic variations in antioxidant activity and silybin content [J]. Russ J Plant Physiol, 2025, 72(2): 45. |
| [28] | Yang M, Yang J, Su L, et al. Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress [J]. Plant Sci, 2019, 289: 110282. |
| [29] | Winkel-Shirley B. Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology [J]. Plant Physiol, 2001, 126(2): 485-493. |
| [30] | Dai H, Xiao CN, Liu HB, et al. Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza Bunge induced by water depletion [J]. J Proteome Res, 2010, 9(3): 1460-1475. |
| [31] | 杨朝结, 张兰, 陈红, 等. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定 [J]. 生物技术通报, 2025, 41(4): 134-144. |
| Yang CJ, Zhang L, Chen H, et al. Functional identification of the transcription factor gene FtbHLH3 in regulating flavonoid biosynthesis in Fagopyrum tataricum [J]. Biotechnol Bull, 2025, 41(4): 134-144. | |
| [32] | Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis [J]. Plant Cell, 1995, 7(7): 1071-1083. |
| [1] | 刘语诗, 李镇, 邹宇琛, 汤维维, 李彬. 药用植物空间代谢组学研究进展[J]. 生物技术通报, 2025, 41(9): 22-31. |
| [2] | 张雅祺, 王芹芹, 沈夏, 李旭苗, 高敏, 李军, 李辰, 王慧. 食管鳞状细胞癌早期进展风险的代谢物预警模型[J]. 生物技术通报, 2025, 41(9): 335-344. |
| [3] | 蒋天威, 马培杰, 李亚娇, 陈才俊, 刘晓霞, 王小利. 二穗短柄草对光周期的代谢响应分析[J]. 生物技术通报, 2025, 41(7): 237-247. |
| [4] | 张越, 毕钰, 慕雪男, 郑子薇, 王志刚, 徐伟慧. 小麦赤霉病拮抗菌JB7的生防特性[J]. 生物技术通报, 2025, 41(7): 261-271. |
| [5] | 杨宗辉, 李利斌, 孟昭娟, 高天, 祝利霞, 杜海梅, 董伟伟, 曹齐卫. 比较转录组学揭示乙烯信号与表观遗传协同调控黄瓜性别决定[J]. 生物技术通报, 2025, 41(12): 139-155. |
| [6] | 罗宜菲, 徐娅, 卢斯琪, 高媛媛, 苟展, 方红, 尚轶, 滕林琳. 致病疫霉侵染前后马铃薯植株挥发性成分分析[J]. 生物技术通报, 2025, 41(11): 247-260. |
| [7] | 韩凯, 周永顺, 张凯月, 王路, 高剑峰, 陈福龙. 三株小球藻抗旱性能评价[J]. 生物技术通报, 2024, 40(8): 244-254. |
| [8] | 王睿, 戚继. 整合组织学图像信息增强空间转录组细胞聚类的分辨率[J]. 生物技术通报, 2024, 40(8): 39-46. |
| [9] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [10] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| [11] | 韩乐乐, 宋文迪, 边嘉珅, 李阳, 杨双胜, 陈紫怡, 李晓薇. 转录组与代谢组联合分析揭示大豆GmERD15c参与盐胁迫下类黄酮的生物合成[J]. 生物技术通报, 2024, 40(10): 243-252. |
| [12] | 姜宇舢, 兰倩, 王芳, 姜亮, 裴成成. 一个影响酪氨酸代谢藜麦突变体的鉴定[J]. 生物技术通报, 2024, 40(10): 253-261. |
| [13] | 何诗瑜, 曾仲大, 李博岩. 空间分辨代谢组学在疾病诊断研究中的应用进展[J]. 生物技术通报, 2024, 40(1): 145-159. |
| [14] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [15] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||