








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 1-15.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0637
• 综述与专论 • 下一篇
收稿日期:2025-06-18
出版日期:2025-12-26
发布日期:2026-01-06
作者简介:方玉洁,女,博士,副教授,研究方向 :油菜遗传育种与分子生物学;E-mail: yjfang@yzu.edu.cn;方玉洁同时为本文
基金资助:
FANG Yu-jie( ), LIU Kuan, CUI Han-bing, WANG You-ping
), LIU Kuan, CUI Han-bing, WANG You-ping
Received:2025-06-18
Published:2025-12-26
Online:2026-01-06
摘要:
SUMO化修饰作为真核生物中一种重要的可逆蛋白翻译后修饰形式,通过调控蛋白质的稳定性、亚细胞定位和活性参与植物的多种生命活动过程。SUMO化修饰过程由一系列酶促级联反应介导,其中SUMO E3连接酶在靶蛋白选择中具有决定性作用。近年研究发现,SUMO E3连接酶通过介导对多类靶蛋白的SUMO化修饰,在植物的生长发育与逆境适应中发挥关键调控作用。本文系统综述了植物中SUMO E3连接酶的结构特征、功能分化及其在多种生理过程中的调控网络,重点梳理其在非生物胁迫响应、生长发育以及免疫应答调控中的分子机制及信号整合途径,旨在为深入解析植物SUMO化修饰的调控网络与生物学功能提供理论参考,并为作物抗性改良与高产育种提供潜在靶点。
方玉洁, 刘宽, 崔寒冰, 王幼平. 植物SUMO E3连接酶研究进展[J]. 生物技术通报, 2025, 41(12): 1-15.
FANG Yu-jie, LIU Kuan, CUI Han-bing, WANG You-ping. Recent Advances in Plant SUMO E3 Ligases[J]. Biotechnology Bulletin, 2025, 41(12): 1-15.
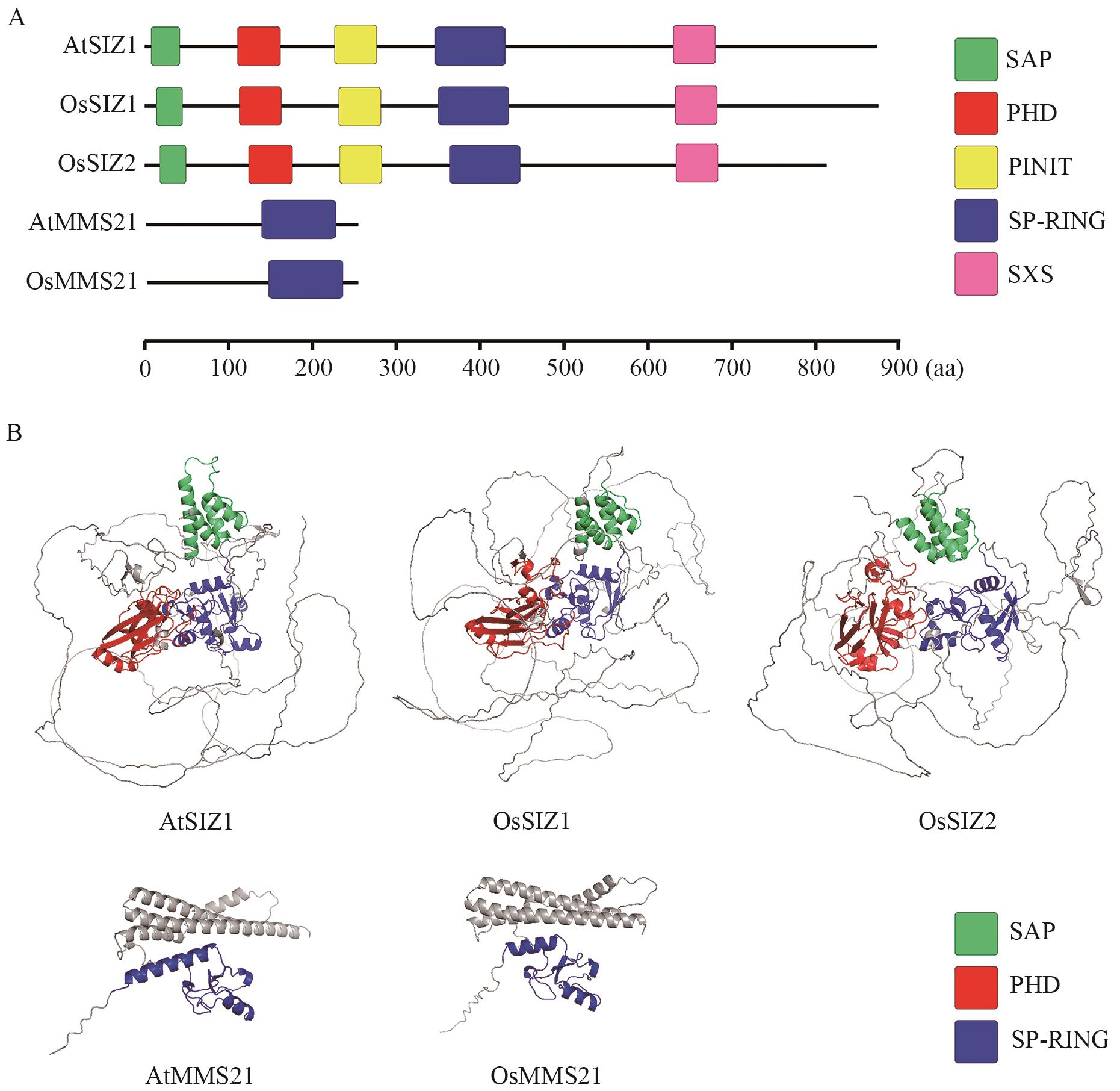
图1 拟南芥与水稻中SUMO E3连接酶的保守结构域A. SUMO E3连接酶的保守结构域示意图;B. SUMO E3连接酶的三级结构预测
Fig. 1 Conserved domains of SUMO E3 ligases in Arabidopsis and riceA. Diagram of conserved domains of SUMO E3 ligases. B. Prediction of the tertiary structure of SUMO E3 ligases
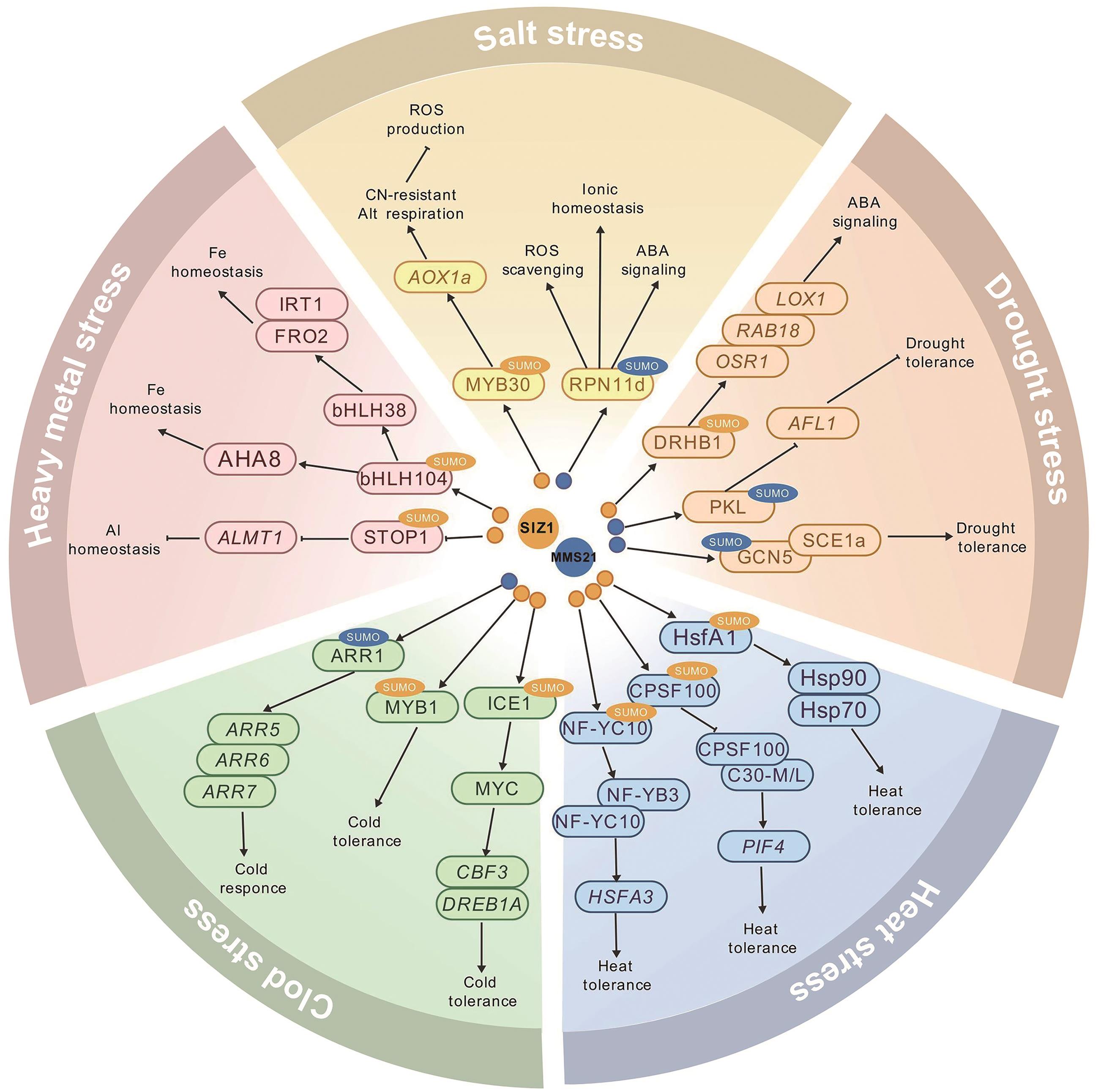
图2 SUMO E3连接酶介导的SUMO化修饰在植物非生物胁迫响应中的调控网络:SIZ1介导的SUMO化;:MMS21介导的SUMO化;→:正调节;:负调节
Fig. 2 The regulatory network of SUMO E3 ligase-mediated sumoylation in plant abiotic stress responses: SIZ1-mediated sumoylation; : MMS21-mediated sumoylation; →: positive regulation; : negative regulation
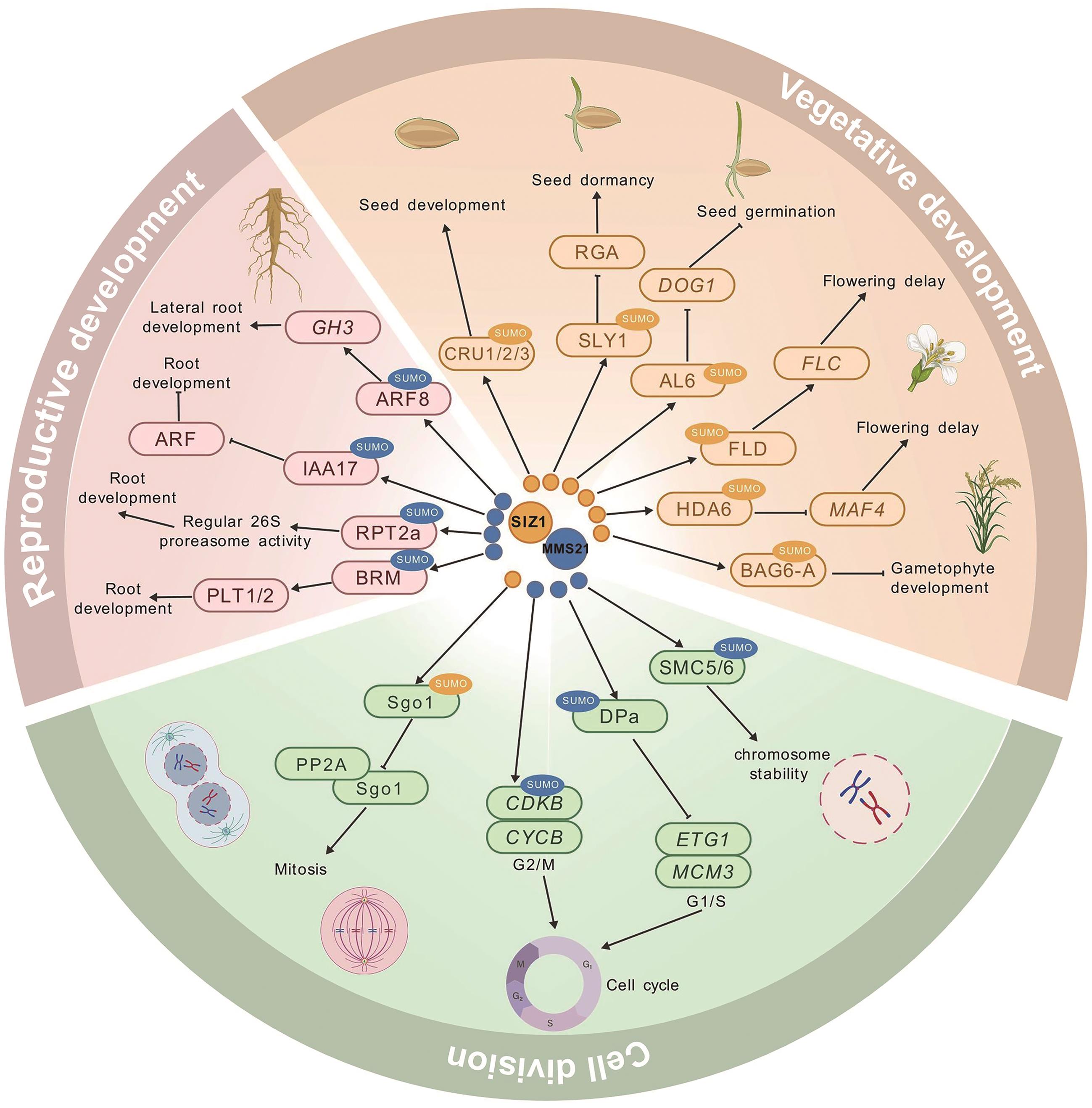
图3 SUMO E3连接酶介导的SUMO化修饰在植物生长发育中的调控网络:SIZ1介导的SUMO化;:MMS21介导的SUMO化;→:正调节;:负调节
Fig. 3 Regulatory network of SUMO E3 ligase-mediated sumoylation in plant growth and development: SIZ1-mediated sumoylation; : MMS21-mediated sumoylation; →: positive regulation; : negative regulation
| [1] | 陈艳梅. 蛋白质翻译后修饰之间的互作关系及其协同调控机理 [J]. 生物技术通报, 2024, 40(2): 1-8. |
| Chen YM. Crosstalk between different post-translational modifications and its regulatory mechanisms in plant growth and development [J]. Biotechnol Bull, 2024, 40(2): 1-8. | |
| [2] | Li ZX, Qin LQ, Wang DJ, et al. Protein post-translational modification in plants: Regulation and beyond [J]. Hortic Plant J, 2025 |
| [3] | Benlloch R, Lois LM. Sumoylation in plants: mechanistic insights and its role in drought stress [J]. J Exp Bot, 2018, 69(19): 4539-4554. |
| [4] | Zhang ZX, Dong YJ, Wang XY, et al. Protein post-translational modifications (PTMs) unlocking resilience to abiotic stress in horticultural crops: a review [J]. Int J Biol Macromol, 2025, 306: 141772. |
| [5] | Elrouby N. Analysis of small ubiquitin-like modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development [J]. Plant Physiol, 2015, 169(2): 1006-1017. |
| [6] | Yau TY, Sander W, Eidson C, et al. SUMO interacting motifs: structure and function [J]. Cells, 2021, 10(11): 2825. |
| [7] | Jmii S, Cappadocia L. Plant SUMO E3 ligases: function, structural organization, and connection with DNA [J]. Front Plant Sci, 2021, 12: 652170. |
| [8] | Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis [J]. Nat Rev Mol Cell Biol, 2009, 10(8): 564-568. |
| [9] | Vierstra RD. The expanding universe of ubiquitin and ubiquitin-like modifiers [J]. Plant Physiol, 2012, 160(1): 2-14. |
| [10] | Bernier-Villamor V, Sampson DA, Matunis MJ, et al. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1 [J]. Cell, 2002, 108(3): 345-356. |
| [11] | Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins [J]. Cell, 2001, 106(6): 735-744. |
| [12] | Yunus AA, Lima CD. Structure of the siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA [J]. Mol Cell, 2009, 35(5): 669-682. |
| [13] | Pichler A, Gast A, Seeler JS, et al. The nucleoporin RanBP2 has SUMO1 E3 ligase activity [J]. Cell, 2002, 108(1): 109-120. |
| [14] | Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3 [J]. Cell, 2003, 113(1): 127-137. |
| [15] | Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses [J]. Proc Natl Acad Sci U S A, 2005, 102(21): 7760-7765. |
| [16] | Huang LX, Yang SG, Zhang SC, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root [J]. Plant J, 2009, 60(4): 666-678. |
| [17] | Cheong MS, Park HC, Bohnert HJ, et al. Structural and functional studies of SIZ1, a PIAS-type SUMO E3 ligase from Arabidopsis [J]. Plant Signal Behav, 2010, 5(5): 567-569. |
| [18] | Park HC, Kim H, Koo SC, et al. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice [J]. Plant Cell Environ, 2010, 33(11): 1923-1934. |
| [19] | Ishida T, Yoshimura M, Miura K, et al. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development [J]. PLoS One, 2012, 7(10): e46897. |
| [20] | Catala R, Ouyang J, Abreu IA, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses [J]. Plant Cell, 2007, 19(9): 2952-2966. |
| [21] | Yoo CY, Miura K, Jin JB, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid [J]. Plant Physiol, 2006, 142(4): 1548-1558. |
| [22] | Zhou LJ, Li YY, Zhang RF, et al. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple [J]. Plant Cell Environ, 2017, 40(10): 2068-2080. |
| [23] | Lee J, Nam J, Park HC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase [J]. Plant J, 2007, 49(1): 79-90. |
| [24] | Miura K, Sato A, Ohta M, et al. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2 [J]. Planta, 2011, 234(6): 1191-1199. |
| [25] | Mishra N, Srivastava AP, Esmaeili N, et al. Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought-, heat-, and salt-tolerance simultaneously [J]. PLoS One, 2018, 13(8): e0201716. |
| [26] | Han GL, Yuan F, Guo JR, et al. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis [J]. Plant Sci, 2019, 285: 55-67. |
| [27] | Gong QY, Li S, Zheng Y, et al. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis [J]. Plant J, 2020, 102(6): 1157-1171. |
| [28] | Lei SK, Wang QZ, Chen Y, et al. Capsicum SIZ1 contributes to ABA-induced SUMOylation in pepper [J]. Plant Sci, 2022, 314: 111099. |
| [29] | Wang SB, Guo JS, Peng YY, et al. GmRPN11d positively regulates plant salinity tolerance by improving protein stability through SUMOylation [J]. Int J Biol Macromol, 2025, 294: 139393. |
| [30] | Joo H, Lim CW, Lee SC. Pepper SUMO E3 ligase CaDSIZ1 enhances drought tolerance by stabilizing the transcription factor CaDRHB1 [J]. New Phytol, 2022, 235(6): 2313-2330. |
| [31] | Zhang SC, Qi YL, Liu M, et al. SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana [J]. J Integr Plant Biol, 2013, 55(1): 83-95. |
| [32] | Castro PH, Couto D, Freitas S, et al. SUMO proteases ULP1c and ULP1d are required for development and osmotic stress responses in Arabidopsis thaliana [J]. Plant Mol Biol, 2016, 92(1-2): 143-159. |
| [33] | Jing YX, Yang ZY, Yang RZ, et al. PKL is stabilized by MMS21 to negatively regulate Arabidopsis drought tolerance through directly repressing AFL1 transcription [J]. New Phytol, 2023, 239(3): 920-935. |
| [34] | Feng TY, Wang YX, Zhang MC, et al. ZmSCE1a positively regulates drought tolerance by enhancing the stability of ZmGCN5 [J]. Plant J, 2024, 120(5): 2101-2112. |
| [35] | Hammoudi V, Fokkens L, Beerens B, et al. The Arabidopsis SUMO E3 ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth [J]. PLoS Genet, 2018, 14(1): e1007157. |
| [36] | Rytz TC, Miller MJ, McLoughlin F, et al. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress [J]. Plant Cell, 2018, 30(5): 1077-1099. |
| [37] | Zhang S, Wang SJ, Lv JL, et al. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato [J]. Plant Cell Physiol, 2018, 59(1): 58-71. |
| [38] | Yu ZB, Wang J, Zhang C, et al. SIZ1-mediated SUMOylation of CPSF100 promotes plant thermomorphogenesis by controlling alternative polyadenylation [J]. Mol Plant, 2024, 17(9): 1392-1406. |
| [39] | Zheng XT, Wang CJ, Lin WX, et al. Importation of chloroplast proteins under heat stress is facilitated by their SUMO conjugations [J]. New Phytol, 2022, 235(1): 173-187. |
| [40] | Huang JW, Huang JJ, Feng QY, et al. SUMOylation facilitates the assembly of a Nuclear Factor-Y complex to enhance thermotolerance in Arabidopsis [J]. J Integr Plant Biol, 2023, 65(3): 692-702. |
| [41] | Kwak JS, Song JT, Seo HS. E3 SUMO ligase SIZ1 splicing variants localize and function according to external conditions [J]. Plant Physiol, 2024, 195(2): 1601-1623. |
| [42] | Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis [J]. Plant Cell, 2007, 19(4): 1403-1414. |
| [43] | Miura K, Ohta M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation [J]. J Plant Physiol, 2010, 167(7): 555-560. |
| [44] | Kim JY, Jang IC, Seo HS. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity [J]. Front Plant Sci, 2016, 7: 1182. |
| [45] | Jiang H, Zhou LJ, Gao HN, et al. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple [J]. Plant Physiol, 2022, 189(4): 2044-2060. |
| [46] | Kang NY, Kim MJ, Jeong S, et al. HIGH PLOIDY2-mediated SUMOylation of transcription factor ARR1 controls two-component signaling in Arabidopsis [J]. Plant Cell, 2024, 36(9): 3521-3542. |
| [47] | Datta M, Kaushik S, Jyoti A, et al. SIZ1-mediated SUMOylation during phosphate homeostasis in plants: Looking beyond the tip of the iceberg [J]. Semin Cell Dev Biol, 2018, 74: 123-132. |
| [48] | Kenji Miura JL. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation [J]. Plant Physiol, 2011, 155(2): 1000-1012. |
| [49] | Pei WX, Jain A, Sun YF, et al. OsSIZ2 exerts regulatory influences on the developmental responses and phosphate homeostasis in rice [J]. Sci Rep, 2017, 7: 12280. |
| [50] | Wang HD, Sun R, Cao Y, et al. OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice [J]. Plant Cell Physiol, 2015, 56(12): 2381-2395. |
| [51] | Zhang RF, Zhou LJ, Li YY, et al. Apple SUMO E3 ligase MdSIZ1 is involved in the response to phosphate deficiency [J]. J Plant Physiol, 2019, 232: 216-225. |
| [52] | Fang Q, Zhang J, Yang DL, et al. The SUMO E3 ligase SIZ1 partially regulates STOP1 SUMOylation and stability in Arabidopsis thaliana [J]. Plant Signal Behav, 2021, 16(5): 1899487. |
| [53] | Xu JM, Zhu JY, Liu JJ, et al. SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis [J]. J Integr Plant Biol, 2021, 63(6): 1147-1160. |
| [54] | Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1 [J]. Nat Commun, 2011, 2: 400. |
| [55] | Kim JY, Park BS, Park SW, et al. Nitrate reductases are relocalized to the nucleus by AtSIZ1 and their levels are negatively regulated by COP1 and ammonium [J]. Int J Mol Sci, 2018, 19(4): 1202. |
| [56] | Park BS, Kim SI, Song JT, et al. Arabidopsis SIZ1 positively regulates alternative respiratory bypass pathways [J]. BMB Rep, 2012, 45(6): 342-347. |
| [57] | Youn K, Jung H, Kim SI, et al. Arabidopsis CMT3 activity is positively regulated by AtSIZ1-mediated sumoylation [J]. Plant Sci, 2015, 239: 209-215. |
| [58] | Castro PH, Verde N, Lourenço T, et al. SIZ1-dependent post-translational modification by SUMO modulates sugar signaling and metabolism in Arabidopsis thaliana [J]. Plant Cell Physiol, 2015, 56(12): 2297-2311. |
| [59] | Castro PH, Verde N, Tavares RM, et al. Sugar signaling regulation by Arabidopsis SIZ1-driven sumoylation is independent of salicylic acid [J]. Plant Signal Behav, 2018, 13(4): e1179417. |
| [60] | Castro PH, Verde N, Azevedo H. Arabidopsis thaliana growth is independently controlled by the SUMO E3 ligase SIZ1 and Hexokinase 1 [J]. MicroPubl Biol, 2020, (2020). |
| [61] | Rawat SS, Sandhya S, Laxmi A. Complex genetic interaction between glucose sensor HXK1 and E3 SUMO ligase SIZ1 in regulating plant morphogenesis [J]. Plant Signal Behav, 2024, 19(1): 2341506. |
| [62] | Chen CC, Chen YY, Tang IC, et al. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance [J]. Plant Physiol, 2011, 156(4): 2225-2234. |
| [63] | Chen CC, Chen YY, Yeh KC. Effect of Cu content on the activity of Cu/ZnSOD1 in the Arabidopsis SUMO E3 ligase siz1 mutant [J]. Plant Signal Behav, 2011, 6(10): 1428-1430. |
| [64] | Chen CC, Chien WF, Lin NC, et al. Alternative functions of Arabidopsis Yellow STRIPE-LIKE3: from metal translocation to pathogen defense [J]. PLoS One, 2014, 9(5): e98008. |
| [65] | Zhan EB, Zhou HP, Li S, et al. OTS1-dependent deSUMOylation increases tolerance to high copper levels in Arabidopsis [J]. J Integr Plant Biol, 2018, 60(4): 310-322. |
| [66] | Zheng Z, Liu D. SIZ1 regulates phosphate deficiency-induced inhibition of primary root growth of Arabidopsis by modulating Fe accumulation and ROS production in its roots [J]. Plant Signal Behav, 2021, 16(10): 1946921. |
| [67] | Zhou LJ, Zhang CL, Zhang RF, et al. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis [J]. Plant Physiol, 2019, 179(1): 88-106. |
| [68] | Kenji Miura JL. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling [J]. Proc Natl Acad Sci U S A, 2009, 106(13): 5418-5423. |
| [69] | Zhang JJ, Lai JB, Wang FG, et al. A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development [J]. Plant Physiol, 2017, 173(3): 1574-1582. |
| [70] | Yu MY, Meng BL, Wang FG, et al. A SUMO ligase AtMMS21 regulates activity of the 26S proteasome in root development [J]. Plant Sci, 2019, 280: 314-320. |
| [71] | Zhang CL, Wang GL, Zhang YL, et al. Apple SUMO E3 ligase MdSIZ1 facilitates SUMOylation of MdARF8 to regulate lateral root formation [J]. New Phytol, 2021, 229(4): 2206-2222. |
| [72] | Jiang JM, Xie Y, Du JJ, et al. A SUMO ligase OsMMS21 regulates rice development and auxin response [J]. J Plant Physiol, 2021, 263: 153447. |
| [73] | Zhang C, Yang Y, Yu ZB, et al. SUMO E3 ligase AtMMS21-dependent SUMOylation of AUXIN/INDOLE-3-ACETIC ACID 17 regulates auxin signaling [J]. Plant Physiol, 2023, 191(3): 1871-1883. |
| [74] | Zhang LE, Han Q, Xiong JW, et al. Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana [J]. Plant Cell Physiol, 2019, 60(10): 2282-2292. |
| [75] | Xiong JW, Yang FB, Wei F, et al. Inhibition of SIZ1-mediated SUMOylation of HOOKLESS1 promotes light-induced apical hook opening in Arabidopsis [J]. Plant Cell, 2023, 35(6): 2027-2043. |
| [76] | Zhao FL, Liu LF, Du JK, et al. BAG6-A from Fragaria viridis pollen modulates gametophyte development in diploid strawberry [J]. Plant Sci, 2023, 330: 111667. |
| [77] | Soo K, Kim SI, Woo P, et al. E3 SUMO ligase AtSIZ1 regulates the cruciferin content of Arabidopsis seeds [J]. Biochem Biophys Res Commun, 2019, 519(4): 761-766. |
| [78] | Kim SI, Park BS, Kim DY, et al. E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development [J]. Biochem J, 2015, 469(2): 299-314. |
| [79] | Kim SI, Kwak JS, Song JT, et al. The E3 SUMO ligase AtSIZ1 functions in seed germination in Arabidopsis [J]. Physiol Plant, 2016, 158(3): 256-271. |
| [80] | Jing H, Liu W, Qu GP, et al. SUMOylation of AL6 regulates seed dormancy and thermoinhibition in Arabidopsis [J]. New Phytol, 2025, 245(3): 1040-1055. |
| [81] | Thangasamy S, Guo CL, Chuang MH, et al. Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence [J]. New Phytol, 2011, 189(3): 869-882. |
| [82] | Liu C, Yu HS, Li LG. SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana [J]. PLoS Genet, 2019, 15(1): e1007928. |
| [83] | Son GH, Park BS, Song JT, et al. FLC-mediated flowering repression is positively regulated by sumoylation [J]. J Exp Bot, 2014, 65(1): 339-351. |
| [84] | Jin JB, Jin YH, Lee J, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure [J]. Plant J, 2008, 53(3): 530-540. |
| [85] | Kwak JS, Son GH, Kim SI, et al. Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes flowering locus C through its E3 ligase activity [J]. Front Plant Sci, 2016, 7: 530. |
| [86] | Gao SJ, Zeng XQ, Wang JH, et al. Arabidopsis SUMO E3 ligase SIZ1 interacts with HDA6 and negatively regulates HDA6 function during flowering [J]. Cells, 2021, 10(11): 3001. |
| [87] | Xu PL, Yuan DK, Liu M, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots [J]. Plant Physiol, 2013, 161(4): 1755-1768. |
| [88] | Liu M, Shi SF, Zhang SC, et al. SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis [J]. BMC Plant Biol, 2014, 14(1): 153. |
| [89] | Zhang JY, Augustine RC, Suzuki M, et al. The SUMO ligase MMS21 profoundly influences maize development through its impact on genome activity and stability [J]. PLoS Genet, 2021, 17(10): e1009830. |
| [90] | Yoshimura M, Ishida T. Generation of viable hypomorphic and null mutant plants via CRISPR-Cas9 targeting mRNA splicing sites [J]. J Plant Res, 2025, 138(1): 189-196. |
| [91] | Liu YY, Lai JB, Yu MY, et al. The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation [J]. Plant Cell, 2016, 28(9): 2225-2237. |
| [92] | Su XB, Wang ML, Schaffner C, et al. SUMOylation stabilizes sister kinetochore biorientation to allow timely anaphase [J]. J Cell Biol, 2021, 220(7): e202005130. |
| [93] | Wang GL, Zhang CL, Huo HQ, et al. The SUMO E3 ligase MdSIZ1 sumoylates a cell number regulator MdCNR8 to control organ size [J]. Front Plant Sci, 2022, 13: 836935. |
| [94] | Yuan MH, Ngou BPM, Ding PT, et al. PTI-ETI crosstalk: an integrative view of plant immunity [J]. Curr Opin Plant Biol, 2021, 62: 102030. |
| [95] | Zhang Y, Zeng LR. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity [J]. Plant Commun, 2020, 1(4): 100041. |
| [96] | Fu ZQ, Dong XN. Systemic acquired resistance: turning local infection into global defense [J]. Annu Rev Plant Biol, 2013, 64: 839-863. |
| [97] | Gou MY, Huang QS, Qian WQ, et al. Sumoylation E3 ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in Arabidopsis [J]. Mol Plant Microbe Interact, 2017, 30(4): 334-342. |
| [98] | Niu D, Lin XL, Kong XX, et al. SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis [J]. Mol Plant, 2019, 12(2): 215-228. |
| [99] | Xie B, Luo MY, Li QY, et al. NUA positively regulates plant immunity by coordination with ESD4 to deSUMOylate TPR1 in Arabidopsis [J]. New Phytol, 2024, 241(1): 363-377. |
| [100] | Liu S, Lenoir CJG, Amaro TMMM, et al. Virulence strategies of an insect herbivore and oomycete plant pathogen converge on host E3 SUMO ligase SIZ1 [J]. New Phytol, 2022, 235(4): 1599-1614. |
| [101] | Hou SG, Liu ZY, Shen HX, et al. Damage-associated molecular pattern-triggered immunity in plants [J]. Front Plant Sci, 2019, 10: 646. |
| [102] | Zhang C, Wu YY, Liu JE, et al. SUMOylation controls peptide processing to generate damage-associated molecular patterns in Arabidopsis [J]. Dev Cell, 2025, 60(5): 696-705.e4. |
| [103] | Orosa B, Yates G, Verma V, et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity [J]. Nat Commun, 2018, 9: 5185. |
| [104] | Shi XX, Du YX, Li SJ, et al. The role of SUMO E3 ligases in signaling pathway of cancer cells [J]. Int J Mol Sci, 2022, 23(7): 3639. |
| [105] | Sang T, Xu YP, Qin GC, et al. Highly sensitive site-specific SUMOylation proteomics in Arabidopsis [J]. Nat Plants, 2024, 10(9): 1330-1342. |
| [106] | Augustine RC, York SL, Rytz TC, et al. Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress [J]. Plant Physiol, 2016, 171(3): 2191-2210. |
| [107] | Clark L, Sue-Ob K, Mukkawar V, et al. Understanding SUMO-mediated adaptive responses in plants to improve crop productivity [J]. Essays Biochem, 2022, 66(2): 155-168. |
| [1] | 王从欢, 伍国强, 魏明. 植物CBL调控逆境胁迫响应的作用机制[J]. 生物技术通报, 2025, 41(7): 1-16. |
| [2] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [3] | 昝舒雯, 谢欢欢, 张宇琴, 王文娟, 张鹏飞, 梁晋军, 温鹏飞. VvAGAMOUS通过VvCRABS CLAW调控葡萄心皮发育[J]. 生物技术通报, 2025, 41(5): 208-217. |
| [4] | 王斌, 林薇, 肖艳辉, 袁晓. 植物富含甘氨酸蛋白家族功能研究进展[J]. 生物技术通报, 2025, 41(2): 1-17. |
| [5] | 吴丁洁, 陈盈盈, 徐静, 刘源, 张航, 李瑞丽. 植物赤霉素氧化酶及其功能研究进展[J]. 生物技术通报, 2024, 40(7): 43-54. |
| [6] | 陈盈盈, 吴丁洁, 刘源, 张航, 刘艳娇, 王晶宇, 李瑞丽. 14-3-3蛋白及其在植物中的功能研究进展[J]. 生物技术通报, 2024, 40(4): 12-22. |
| [7] | 付威, 韦素云, 陈赢男. 植物生长发育动态QTL解析研究进展[J]. 生物技术通报, 2024, 40(2): 9-19. |
| [8] | 侯鹰翔, 费思恬, 宋松泉, 罗勇, 张超. 水稻MADS-box家族研究进展[J]. 生物技术通报, 2024, 40(11): 103-112. |
| [9] | 陈梦娇, 李洋洋, 邬倩. 谷氨酸受体蛋白调控植物生长与胁迫应答的研究进展[J]. 生物技术通报, 2024, 40(10): 62-75. |
| [10] | 毕芳玲, 赵爽, 栗斌, 李爱芹, 张建恒, 何培民. 共附生菌对绿潮浒苔作用的研究进展及应用[J]. 生物技术通报, 2024, 40(1): 32-44. |
| [11] | 胡海琳, 徐黎, 李晓旭, 王晨璨, 梅曼, 丁文静, 赵媛媛. 小肽激素调控植物生长发育及逆境生理研究进展[J]. 生物技术通报, 2023, 39(7): 13-25. |
| [12] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [13] | 薛皦, 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋. 植物基因上游开放阅读框的研究进展[J]. 生物技术通报, 2023, 39(4): 157-165. |
| [14] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [15] | 桑田, 王鹏程. 植物SUMO化修饰研究进展[J]. 生物技术通报, 2023, 39(3): 1-12. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||