








生物技术通报 ›› 2021, Vol. 37 ›› Issue (8): 195-202.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1361
收稿日期:2020-11-05
出版日期:2021-08-26
发布日期:2021-09-10
作者简介:范晨龙,男,硕士,研究方向:水产经济动物病害防治;E-mail: 基金资助:Received:2020-11-05
Published:2021-08-26
Online:2021-09-10
摘要:
旨在研究溶藻弧菌(Vibrio alginolyticus)去乙酰化酶cobB基因的功能。进行了cobB基因的克隆,生物信息学分析,CobB蛋白质诱导表达、纯化及功能研究。CobB蛋白质的相对分子质量为27.09750 kD,理论等电点为5.15,化学式为C1186H1865N339O369S10,亲水性平均系数(GRAVY):-0.409,为亲水蛋白。CobB与同弧菌属的魔鬼弧菌(Vibrio diabolicus),哈维弧菌(Vibrio harveyi),霍乱弧菌(Vibrio cholerae)和副溶血性弧菌(Vibrio parahaemolyticus)具有高度同源性,在革兰氏阴性菌中相对保守;融合蛋白大小约为53 kD,去除标签后CobB蛋白分子量约为27 kD;CobB蛋白最适表达条件为37℃,0.4 mmol /L IPTG下诱导4 h。功能分析表明,CobB对乙酰化蛋白具有去乙酰化作用。
范晨龙, 丁燏. 溶藻弧菌去乙酰化酶基因cobB克隆及其功能验证[J]. 生物技术通报, 2021, 37(8): 195-202.
FAN Chen-long, DING Yu. Molecular Cloning and Functional Verification of Histone Deacetylase Gene cobB in Vibrio alginolyticus[J]. Biotechnology Bulletin, 2021, 37(8): 195-202.
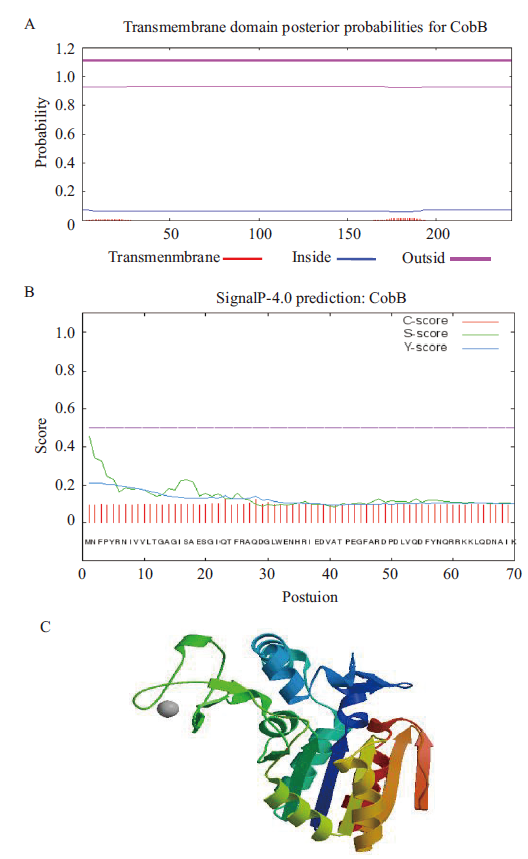
图2 cobB 序列生物信息学分析 A:信号肽预测;B:跨膜结构域预测;C:蛋白质三级结构预测
Fig.2 Bioinformatics analysis of cobB sequences A:Signal peptide prediction. B:Transmembrane domain prediction.C:Protein tertiary structure prediction
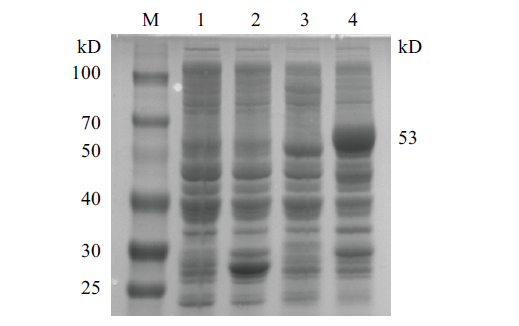
图4 CobB蛋白表达观测实验凝胶电泳示意图 M:蛋白质 marker;1:pGEX-6P-1未诱导;2:pGEX-6P-1诱导;3:pGEX-6P-1-cobB 未诱导;4:pGEX-6P-1-cobB 诱导
Fig.4 Schematic diagram of gel electrophoresis of CobB protein expression M:Protein marker;1:negative control without IPTG;2:negative control with IPTG;3:pGEX-6p-cobB without IPTG;4:pGEX-6p-cobB with IPTG
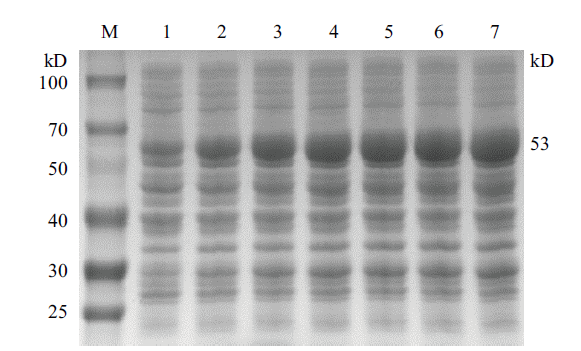
图5 不同诱导时间对 CobB 蛋白表达影响凝胶电泳示意图 M:蛋白质 marker;1-7:诱导时间分别为1 h、2 h、3 h、4 h、5 h、6 h、7 h
Fig.5 Schematic diagram of gel electrophoresis of different induction time on CobB protein expression M:Protein marker;1-7:The induction time is 1 h、2 h、3 h、4 h、5 h、6 h、7 h
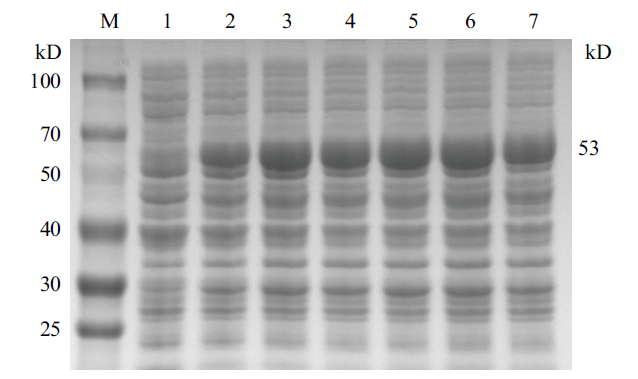
图6 不同IPTG浓度对 CobB 蛋白表达影响凝胶电泳示意图 M:蛋白质 marker;1-7:IPTG浓度分别为 0.1%、0.2%、0.4%、0.6%、0.8%、1% 和 2%
Fig.6 Schematic diagram of gel electrophoresis of different IPTG concentrations on CobB protein expression M:Protein marker;1-7:the IPTG concentrations are 0.1%、0.2%、0.4%、0.6%、0.8%、1% and 2%

图7 不同温度对 CobB 蛋白表达影响凝胶电泳示意图 M:蛋白质marker;1:28℃全菌蛋白;2:28℃破碎后上清;3:28℃破碎后沉淀;4:37℃全菌蛋白;5:37℃破碎后上清;6:37℃破碎后沉淀
Fig.7 Schematic diagram of gel electrophoresis of different temperatures on CobB protein expression M:Protein maker;1:28℃ bacteria protein;2:28℃ supernatant protein;3:28℃ precipitation protein;4:37℃ bacteria protein;5:37℃ supernatant protein;6:37℃ precipitation protein
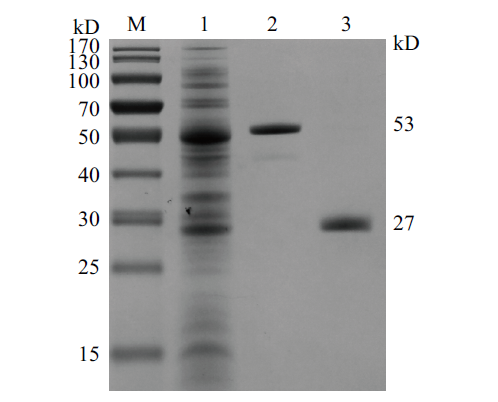
图8 CobB 蛋白纯化酶切凝胶电泳示意图 M:蛋白质 Marker;1:全菌蛋白;2:CobB融合蛋白纯化结果;3:CobB融合蛋白酶切结果
Fig.8 Schematic diagram of digestion gel electrophoresis of CobB protein purification M:Protein molecular standard;1:Bacteria protein;2:Results of CobB fusion protein purification;3:Results of CobB fusion protease cleavage results
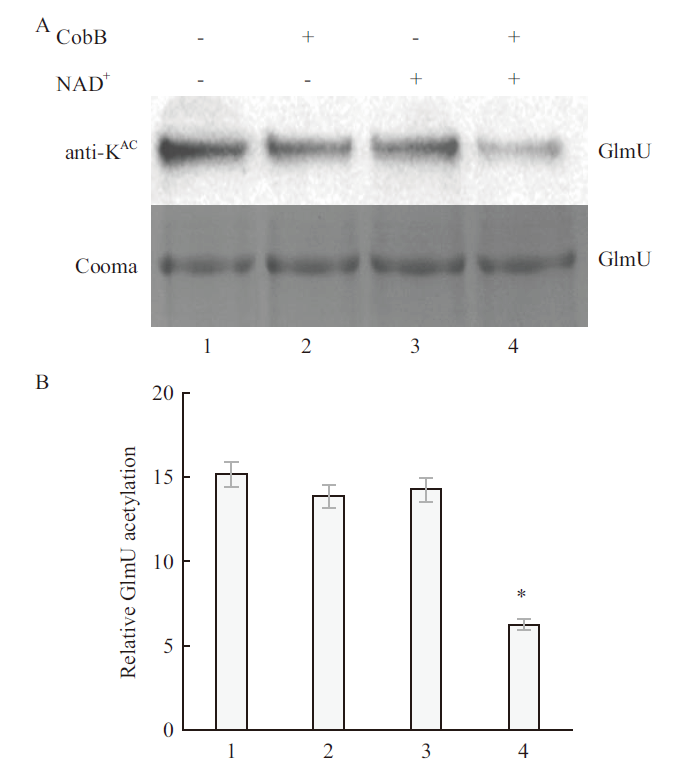
图9 CobB 去乙酰化 GlmU 蛋白的 Western 印迹 A:乙酰赖氨酸特异性抗体的 Western 印迹及 SDS-PAGE;B:Western 印迹定量的 GlmU 乙酰化水平。1:无CobB蛋白,无NAD +组;2:有CobB蛋白,无NAD+组;3:无CobB蛋白,有NAD +组;4:有CobB蛋白,有NAD +组
Fig.9 Western blot of CobB deacetylated GlmU protein A:Western blot of acetyllysine-specific antibodies and SDS-PAGE;B:GlmU acetylation level quantified by Western blot. 1:No CobB protein,no NAD+ group;2:CobB protein,no NAD + group;3:no CobB protein,NAD+ group;4:CobB protein,NAD+ group
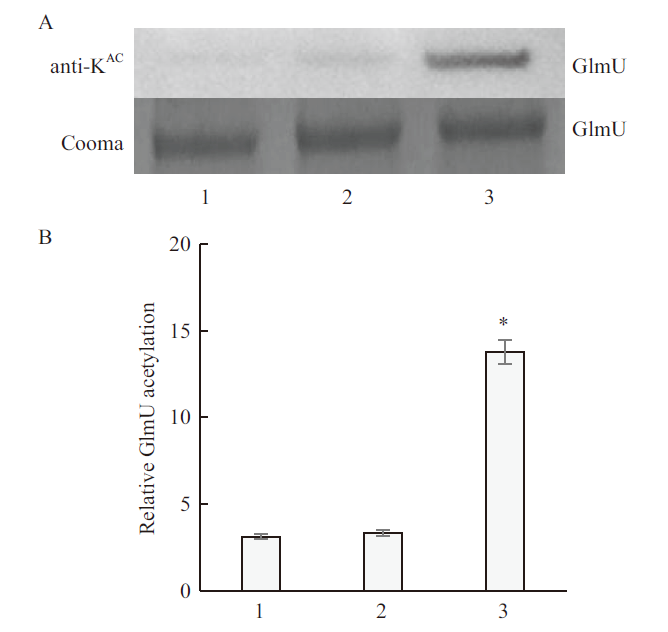
图10 CobB融合蛋白及去除标签的CobB蛋白去乙酰化GlmU蛋白的Western 印迹 A:乙酰赖氨酸特异性抗体的Western印迹及SDS-PAGE;B:Western印迹定量的GlmU乙酰化水平。1:CobB 融合蛋白组;2:CobB蛋白组;3.GST蛋白组
Fig.10 Western blot of CobB fusion protein and CobB protein deacetylated GlmU protein A:Western blot of acetyllysine-specific antibodies and SDS-PAGE;B:GlmU acetylation level quantified by Western blot. 1:CobB fusion protein group;2:CobB protein group;3:GST protein group
| [1] | 常云胜. 溶藻弧菌Ⅲ型分泌系统vscB基因的功能研究及vscD基因的克隆表达[D]. 湛江:广东海洋大学, 2018. |
| Chang YS. Cloning and expression of vscD gene and study on the function of vscB gene in type III secretion system of Vibrio alginolyticus[D]. Zhanjiang:Guangdong Ocean University, 2018. | |
| [2] |
Minguez P, Parcaet L, Diella F, et al. Deciphering a global network of functionally associated post-translational modifications[J]. Molecular Systems Biology, 2012, 8:599.
doi: 10.1038/msb.2012.31 pmid: 22806145 |
| [3] |
Ramponi G, Manao G, Camici G, et al. Nonenzymic acetylation of histones with acetyl phosphate and acetyl adenylate[J]. Biochemistry, 1975, 14(12):2681-2685.
pmid: 238571 |
| [4] |
Zhang L, Yao WL, Xia J, et al. Glucagon-induced acetylation of energy-sensing factors in control of hepatic metabolism[J]. International Journal of Molecular Sciences, 2019, 20:8.
doi: 10.3390/ijms20010008 URL |
| [5] |
Santos JD, Zuma AA, Vitorino FND, et al. Trichostatin A induces Trypanosoma cruzi histone and tubulin acetylation:effects on cell division and microtubule cytoskeleton remodelling[J]. Parasitology, 2019, 146(4):543-552.
doi: 10.1017/S0031182018001828 URL |
| [6] | Zhao GY, Wang H, Xu CZ, et al. SIRT6 delays cellular senescence by promoting p27Kip1 ubiquitin-proteasome degradation[J]. Aging-US, 2016, 8(10):2308-2323. |
| [7] |
Chuang C, Lin SH, Huang F, et al. Acetylation of RNA processing proteins and cell cycle proteins in mitosis[J]. Journal of Proteome Research, 2010, 9(9):4554-4564.
doi: 10.1021/pr100281h pmid: 20812760 |
| [8] |
Jiang W, Jiang P, Yang R, et al. Functional role of SIRT1-induced HMGB1 expression and acetylation in migration, invasion and angiogenesis of ovarian cancer[J]. European Review for Medical and Pharmacological Sciences, 2018, 22(14):4431-4439.
doi: 15494 pmid: 30058682 |
| [9] |
Ellinger J, Schneider AC, Bachmann A, et al. Evaluation of global histone acetylation levels in bladder cancer patients[J]. Anticancer Research, 2016, 36(8):3961-3964.
pmid: 27466500 |
| [10] |
Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins[J]. Biochemical and Biophysical Research Communications, 2000, 273(2):793-798.
pmid: 10873683 |
| [11] |
Tsang AW, Escalante-Semerena JC. CobB, a new member of the sir2 family of eucaryotic regulatory proteins, Is required to compensate for the lack of nicotinate mononucleotide:5, 6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynjournal in Salmonella typhimurium LT2[J]. Journal of Biological Chemistry, 1998, 273(48):31788-31794.
pmid: 9822644 |
| [12] |
Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene:Sir2- like proteins(sirtuins)metabolize NAD and may have protein ADP-ribosyltransferase activity[J]. Biochemical and Biophysical Research Communications, 1999, 260(1):273-279.
pmid: 10381378 |
| [13] |
Tanny JC, Dowd GJ, Huang J, et al. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing[J]. Cell, 1999, 99(7):735-745.
pmid: 10619427 |
| [14] |
Zhao KH, Chai XM, Marmorstein R, et al. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli[J]. Journal of Molecular Biology, 2004, 337(3):731-741.
doi: 10.1016/j.jmb.2004.01.060 URL |
| [15] | Gardner JG, Escalante-Semerena JC. In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene(formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase[J]. Journal of Acteriology, 2009, 191(6):1749-1755. |
| [16] |
Crosby HA, Heiniger EK, Harwood CS, et al. Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris[J]. Molecular Microbiology, 2010, 76(4):874-888.
doi: 10.1111/j.1365-2958.2010.07127.x pmid: 20345662 |
| [17] |
Karel M, Jurgen F, Eva K, et al. CobB1 deacetylase activity in Streptomyces coelicolor[J]. Biochemistry and Cell Biology, 2012, 90(2):179-187.
doi: 10.1139/o11-086 URL |
| [18] |
Starai VJ, Celic I, Cole RN, et al. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine[J]. Science, 2002, 298(5602):2390-2392.
pmid: 12493915 |
| [19] |
Kremer M, Kuhlmann N, Lechner M, et al. Comment on ‘YcgC represents a new protein deacetylase family in prokaryotes’[J]. eLife, 2018, 7:e37798.
doi: 10.7554/eLife.37798 URL |
| [20] |
AbouElfetouh A, Kuhn ML, Hu LI, et al. The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites[J]. Microbiology Open, 2015, 4(1):66-83.
doi: 10.1002/mbo3.2015.4.issue-1 URL |
| [21] | 谷晶. 2013年湖北省暨武汉微生物学会会员代表大会暨学术年会论文摘要集[C]. 武汉: 2013. |
| Gu J. 2013 Hubei Province and Wuhan society of microbiology member representative conference and academic annual conference abstracts[C]. Wuhan: 2013. | |
| [22] | 张群芳. 赖氨酸乙酰化调节大肠杆菌NhoA蛋白活性的研究[D]. 武汉:华中农业大学, 2013. |
| Zhang QF. Reversible lysine acetylation regulates the activity of E. coli N-hydroxyarylamine O-acetyltransferase[D]. Wuhan:Huazhong Agricultural University, 2013. | |
| [23] | 孙曼銮. 乙酰化调节大肠杆菌S-腺苷甲硫氨酸合成酶活性的机制研究[D]. 中国科学院研究生院, 2016. |
| Sun ML. Acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase[D]. Wuhan:University of Chinese Academy of Sciences, 2016. | |
| [24] |
Sun ML, Guo HS, Lu GL, et al. Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase[J]. Acta Biochimica et Biophysica Sinica, 2016, 48(8):723-731.
doi: 10.1093/abbs/gmw066 URL |
| [25] |
Zhou QX, Zhou YN, Jin DJ, et al. Deacetylation of topoisomerase I is an important physiological function of E-coli CobB[J]. Nucleic Acids Research, 2017, 45(9):5349-5358.
doi: 10.1093/nar/gkx250 URL |
| [26] | Xu ZW, Zhang HN, Zhang XR, et al. Interplay between the bacterial protein deacetylase CobB and the second messenger c-di-GMP[J]. EMBO Journal, 2019, 38:18. |
| [27] |
Venkat S, Gregory C, Gan QL, et al. Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli[J]. Chembiochem, 2017, 18(19):1928-1934.
doi: 10.1002/cbic.v18.19 URL |
| [28] | Venkat S, Chen H, McGuire P, et al. Characterizing lysine acetylation of Escherichia coli type II citrate synthase[J]. FEBS J, 2019, 286(14):2799-2808. |
| [29] | Ren J, Sang Y, Lu J, et al. Protein acetylation and its role in bacterial virulence[J]. Trends Microbiol, 2017, 9, 25(9):768-779. |
| [1] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [2] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [3] | 曾福源, 苏泽辉, 周诗慧, 谢妙, 庞欢瑛. 溶藻弧菌PEPCK蛋白原核表达及其乙酰化、琥珀酰化修饰的鉴定[J]. 生物技术通报, 2021, 37(5): 84-91. |
| [4] | 代文双, 刘会云, 杜庆国, 邹枨, 王轲. 组蛋白去乙酰化酶抑制剂(HDACi)对小麦基因编辑效率的影响及转录组学分析[J]. 生物技术通报, 2021, 37(1): 2-14. |
| [5] | 杨世全, 彭丹, 费文杰, 杨丰, 屈高毅, 唐威威, 欧剑萍, 邓湘雯, 周波. 杉木ClKptA/Tpt1基因的克隆及其表达特性分析[J]. 生物技术通报, 2020, 36(8): 15-22. |
| [6] | 闵琪, 高子涵, 姚银, 张华山, 熊海容, 张莉. 共表达HAC1和分子伴侣基因对甘露聚糖酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2020, 36(5): 159-168. |
| [7] | 徐洲, 范晨龙, 丁燏. 溶藻弧菌PepA蛋白原核表达载体的构建及其乙酰化鉴定[J]. 生物技术通报, 2020, 36(12): 75-81. |
| [8] | 钟李婷, 陈秀珍, 唐云, 李俊仁, 王小兵, 刘彦婷, 周璇璇, 詹若挺, 陈立凯. 广藿香FPPS重组蛋白表达及互作蛋白筛选分析[J]. 生物技术通报, 2019, 35(12): 10-15. |
| [9] | 高庆华, 董聪, 王玥, 胡美荣, 王庆庆, 王云鹏, 罗同阳, 刘蕾. 共表达分子伴侣PDI和Ero1对葡萄糖氧化酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2018, 34(7): 174-179. |
| [10] | 万青青,李洁,曾钰,张明明,赵心清,白凤武. MRP8过表达促进重组酿酒酵母外切纤维素酶生产[J]. 生物技术通报, 2018, 34(5): 94-100. |
| [11] | 周钦茂, 谢燕纯, 陈彦梅, 张扬, 柯德森, 陈琼华. 一株高产琼胶酶海洋细菌的筛选与鉴定[J]. 生物技术通报, 2018, 34(12): 140-146. |
| [12] | 刘怡君, 贾宇坤, 王玲芳, 刘虹杏, 杨仙玉. 中华大蟾蜍EDF-1重组蛋白的原核表达、纯化及抗血清的制备[J]. 生物技术通报, 2018, 34(10): 129-134. |
| [13] | 刘雁霞,李少杰,樊振川. 粗糙脉孢菌ERG-11蛋白抗原的原核表达、纯化及其多克隆抗体制备[J]. 生物技术通报, 2017, 33(9): 216-222. |
| [14] | 李玉锋,戴习林,袁新程,周迅,胡彦杰,丁福江. 四个罗氏沼虾抗病选择系抗病力比较分析[J]. 生物技术通报, 2017, 33(7): 203-209. |
| [15] | 赵书梅, 王霖慧, 唐嘉琦, 赵静宜. 溶藻弧菌中酯酶基因的克隆表达及其酶学性质研究[J]. 生物技术通报, 2017, 33(6): 190-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
