








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 247-257.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0217
曹修凯1( ), 王珊2, 葛玲2, 张卫博2, 孙伟1,2(
), 王珊2, 葛玲2, 张卫博2, 孙伟1,2( )
)
收稿日期:2021-02-25
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:曹修凯,男,博士,助理研究员,研究方向:绵羊遗传改良;E-mail: 基金资助:
CAO Xiu-kai1( ), WANG Shan2, GE Ling2, ZHANG Wei-bo2, SUN Wei1,2(
), WANG Shan2, GE Ling2, ZHANG Wei-bo2, SUN Wei1,2( )
)
Received:2021-02-25
Published:2022-01-26
Online:2022-02-22
摘要:
染色体外环形DNA(extrachromosomal circular DNA,eccDNA)指来源于基因组DNA并游离于染色体之外的双链环状DNA分子,它在真核生物中普遍存在。eccDNA在数目、序列长度和基因组来源上存在很大差异,其功能也不尽相同,包括端粒可变延长、rDNA拷贝数维持、衰老、耐药性和肿瘤发生等,eccDNA与畜禽表型相关性已有报道。本文综述了eccDNA的分类、产生机制、功能研究和鉴定方法等,并就eccDNA在动物育种中的应用进行阐述。
曹修凯, 王珊, 葛玲, 张卫博, 孙伟. 染色体外环形DNA研究进展及其在畜禽育种中的应用[J]. 生物技术通报, 2022, 38(1): 247-257.
CAO Xiu-kai, WANG Shan, GE Ling, ZHANG Wei-bo, SUN Wei. Advances in Extrachromosomal Circular DNA and Their Application in Domestic Animal Breeding[J]. Biotechnology Bulletin, 2022, 38(1): 247-257.
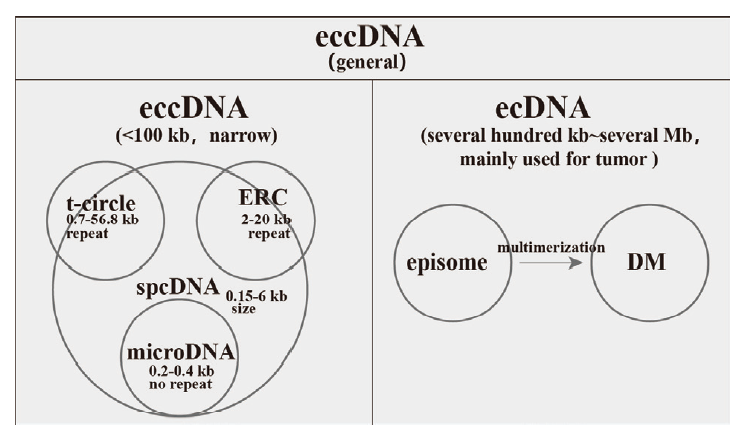
图1 eccDNA的分类 狭义eccDNA小于100 kb,按来源分为t-circle、ERC和microDNA,而spcDNA是早期研究eccDNA提出的概念,它强调的是大小而非序列特征;数百kb到数Mb的eccDNA在癌症中比较常见,命名为ecDNA
Fig.1 eccDNA classification eccDNA in narrow sense are < 100 kb,including t-circle,ERC and microDNA according to their origins. Notably,spcDNA is a concept in the early stage of studying eccDNA,it emphasizes their sizes rather than their sequence features. eccDNA with size ranging from several hundred kilobases to several megabases are common in tumor and termed as ecDNA
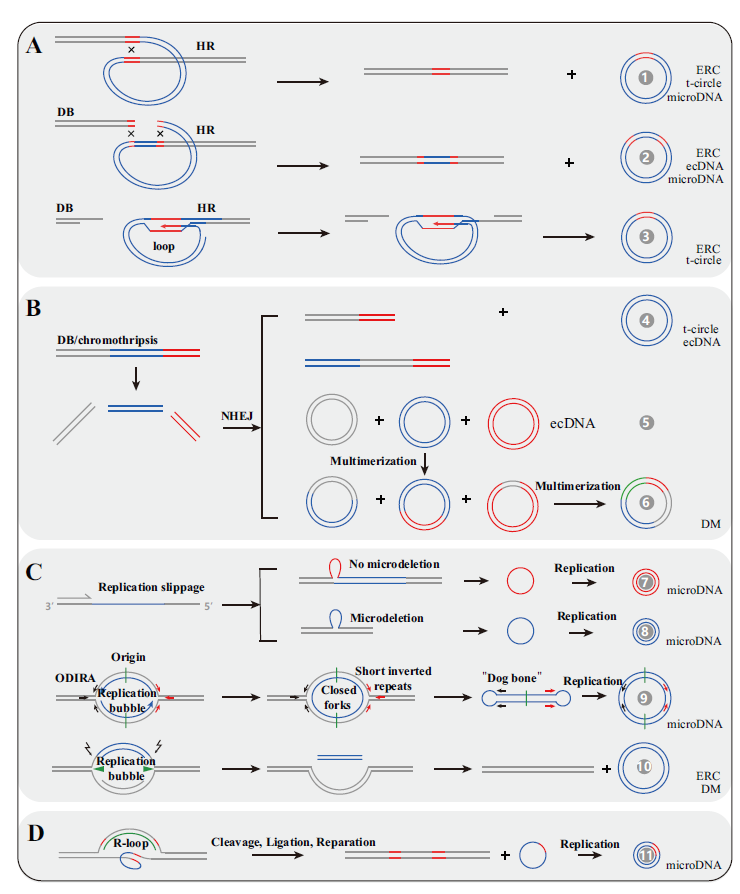
图2 形成eccDNA的潜在机制 A:基于同源重组;B:基于非同源末端连接;C:基于DNA复制;D:基于转录形成的R-loop。形成eccDNA的潜在机制共有11种,参考文献见表1
Fig.2 Possible mechanisms of forming eccDNA A:Formed by HR. B:Formed by NHEJ. C:Formed by DNA replication. D:Formed by transcription mediated by R-loop. There are 11 kinds of potential mechanisms for eccDNA formation and their detail information can be found in references listed in Table 1
| 机制编号 Mechanism No. | eccDNA类型eccDNA type | 时间/类型 Year/Type | 参考文献 Reference |
|---|---|---|---|
| 1 | ERC | 2019,Review | [ |
| 2019,Article | [ | ||
| t-circle | 2004,Review | [ | |
| 2019,Article | [ | ||
| microDNA | 2015,Article | [ | |
| 2019,Article | [ | ||
| 2 | ERC | 1999,Article | [ |
| ecDNA | 2010,Article | [ | |
| microDNA | 2018,Review | [ | |
| 3 | ERC | 2019,Article | [ |
| t-circle | 2004,Review | [ | |
| 2019,Article | [ | ||
| 4 | t-circle | 2004,Review | [ |
| ecDNA | 2020,Review | [ | |
| 2020,Review | [ | ||
| 5 | ecDNA | 2020,Review | [ |
| 2020,Review | [ | ||
| 2020,Review | [ | ||
| 6 | DM | 2020,Review | [ |
| 2020,Review | [ | ||
| 2020,Review | [ | ||
| 7 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 8 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 9 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 10 | ERC | 2018,Article | [ |
| DM | 2004,Article | [ | |
| 2020,Review | [ | ||
| 11 | microDNA | 2018,Review | [ |
| 2020,Review | [ |
表1 形成eccDNA的11种潜在机制对应参考文献
Table 1 Corresponding references of 11 kinds of potential mechanisms for eccDNA formation
| 机制编号 Mechanism No. | eccDNA类型eccDNA type | 时间/类型 Year/Type | 参考文献 Reference |
|---|---|---|---|
| 1 | ERC | 2019,Review | [ |
| 2019,Article | [ | ||
| t-circle | 2004,Review | [ | |
| 2019,Article | [ | ||
| microDNA | 2015,Article | [ | |
| 2019,Article | [ | ||
| 2 | ERC | 1999,Article | [ |
| ecDNA | 2010,Article | [ | |
| microDNA | 2018,Review | [ | |
| 3 | ERC | 2019,Article | [ |
| t-circle | 2004,Review | [ | |
| 2019,Article | [ | ||
| 4 | t-circle | 2004,Review | [ |
| ecDNA | 2020,Review | [ | |
| 2020,Review | [ | ||
| 5 | ecDNA | 2020,Review | [ |
| 2020,Review | [ | ||
| 2020,Review | [ | ||
| 6 | DM | 2020,Review | [ |
| 2020,Review | [ | ||
| 2020,Review | [ | ||
| 7 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 8 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 9 | microDNA | 2015,Article | [ |
| 2018,Review | [ | ||
| 10 | ERC | 2018,Article | [ |
| DM | 2004,Article | [ | |
| 2020,Review | [ | ||
| 11 | microDNA | 2018,Review | [ |
| 2020,Review | [ |
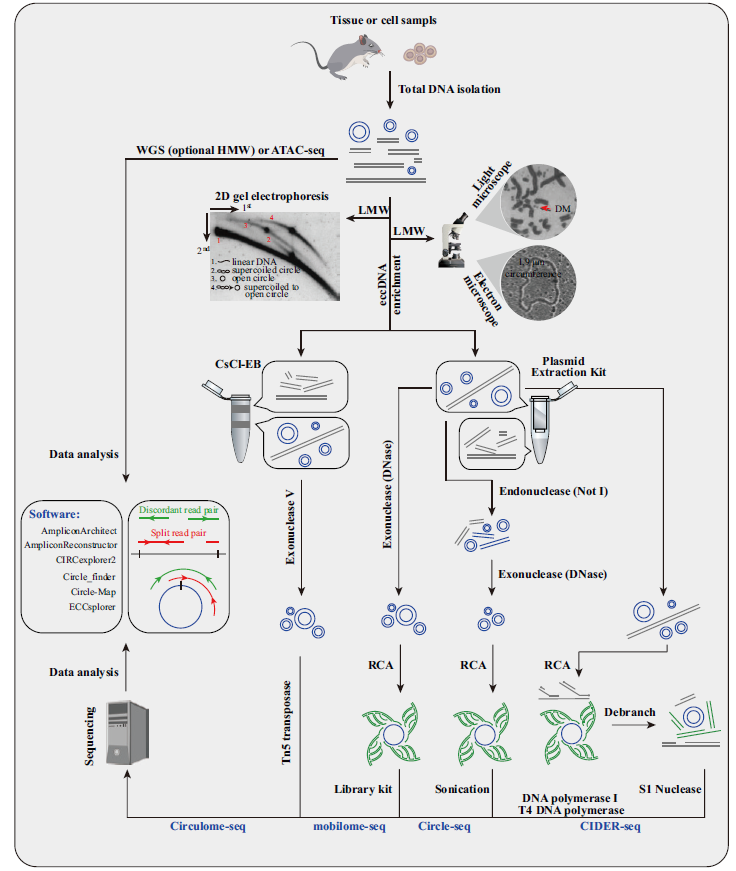
图4 eccDNA的鉴定方法 样品总DNA提取之后,可直接进行显微观测,也可简单富集低分子量(low molecular weight,LMW)DNA后进行显微观测和2D电泳[10,35,88]。高通量测序文库构建可采用CsCl-EB或质粒提取试剂盒富集期望大小的eccDNA,经滚环扩增(rolling circle amplification,RCA)或Tn5处理后,进行高通量测序,利用split read pair和discordant read pair鉴定eccDNA。基因组WGS和ATAC-seq数据也可用于鉴定eccDNA
Fig.4 Methods for eccDNA identification Microscope and 2D electrophoresis are used for eccDNA identification in total DNA or after enrichment of low molecular weight(LMW)DNA[10,35,88]. High-throughput sequencing library is constructed by:eccDNA of expected sizes are enriched by CsCl-EB or plasmid extraction kit,and then high-throughput sequencing is conducted after rolling circle amplification(RCA)or Tn5 treatment. Split and discordant read pairs are used for eccDNA identification. Genome WGS and ATAC-seq data are also for eccDNA identification
| [1] |
Durkin K, Coppieters W, Drögemüller C, et al. Serial translocation by means of circular intermediates underlies colour sidedness in cattle[J]. Nature, 2012, 482(7383):81-84.
doi: 10.1038/nature10757 URL |
| [2] |
Møller HD, Ramos-Madrigal J, Prada-Luengo I, et al. Near-random distribution of chromosome-derived circular DNA in the condensed genome of pigeons and the larger, more repeat-rich human genome[J]. Genome Biol Evol, 2020, 12(1):3762-3777.
doi: 10.1093/gbe/evz281 URL |
| [3] |
Koo DH, Molin WT, Saski CA, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri[J]. PNAS, 2018, 115(13):3332-3337.
doi: 10.1073/pnas.1719354115 URL |
| [4] |
Molin WT, Yaguchi A, Blenner M, et al. The EccDNA replicon:a heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri[J]. Plant Cell, 2020, 32(7):2132-2140.
doi: 10.1105/tpc.20.00099 URL |
| [5] |
Dahm R. Friedrich Miescher and the discovery of DNA[J]. Dev Biol, 2005, 278(2):274-288.
doi: 10.1016/j.ydbio.2004.11.028 URL |
| [6] |
Watson JD, Crick FHC. The structure of dna[J]. Cold Spring Harb Symp Quant Biol, 1953, 18:123-131.
doi: 10.1101/SQB.1953.018.01.020 URL |
| [7] |
Hotta Y, Bassel A. Molecular size and circularity of DNA in cells of mammals and higher plants[J]. PNAS, 1965, 53:356-362.
doi: 10.1073/pnas.53.2.356 URL |
| [8] |
Yamagishi H, Kunisada T, Tsuda T. Small circular DNA complexes in eucaryotic cells[J]. Plasmid, 1982, 8(3):299-306.
pmid: 6294712 |
| [9] |
Cox D, Yuncken C, Spriggs AI. Minute chromatin bodies in malignant tumours of childhood[J]. Lancet, 1965, 1(7402):55-58.
doi: 10.1016/S0140-6736(02)95344-4 URL |
| [10] |
Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA:the closed circular DNA in HeLa cells[J]. PNAS, 1967, 57(5):1514-1521.
pmid: 5231757 |
| [11] |
Smith CA, Vinograd J. Small polydisperse circular DNA of HeLa cells[J]. J Mol Biol, 1972, 69(2):163-178.
pmid: 5070865 |
| [12] |
Stanfield S, Helinski DR. Small circular DNA in Drosophila melanogaster[J]. Cell, 1976, 9(2):333-345.
pmid: 824055 |
| [13] |
DeLap RJ, Rush MG, Zouzias D, et al. Isolation and preliminary characterization of the small circular DNA present in African green monkey kidney(BSC-1)cells[J]. Plasmid, 1978, 1(4):508-521.
pmid: 107539 |
| [14] |
DeLap RJ, Rush MG. Change in quantity and size distribution of small circular DNAs during development of chicken Bursa[J]. PNAS, 1978, 75(12):5855-5859.
pmid: 282606 |
| [15] |
Krolewski JJ, Bertelsen AH, Humayun MZ, et al. Members of the Alu family of interspersed, repetitive DNA sequences are in the small circular DNA population of monkey cells grown in culture[J]. J Mol Biol, 1982, 154(3):399-415.
pmid: 7077666 |
| [16] |
Bertelsen AH, Humayun MZ, Karfopoulos SG, et al. Molecular characterization of small polydisperse circular deoxyribonucleic acid from an African green monkey cell line[J]. Biochemistry, 1982, 21(9):2076-2085.
pmid: 7093232 |
| [17] |
Kazazian HH, Wong C, Youssoufian H, et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man[J]. Nature, 1988, 332(6160):164-166.
doi: 10.1038/332164a0 URL |
| [18] |
Schindler CW, Rush MG. The KpnI family of long interspersed nucleotide sequences is present on discrete sizes of circular DNA in monkey(BSC-1)cells[J]. J Mol Biol, 1985, 181(2):161-173.
pmid: 2984431 |
| [19] |
Kunisada T, Yamagishi H. Sequence organization of repetitive sequences enriched in small polydisperse circular DNAs from HeLa cells[J]. J Mol Biol, 1987, 198(4):557-565.
pmid: 3430621 |
| [20] |
Misra R, Matera AG, Schmid CW, et al. Recombination mediates production of an extrachromosomal circular DNA containing a transposon-like human element, THE-1[J]. Nucleic Acids Res, 1989, 17(20):8327-8341.
pmid: 2478961 |
| [21] |
Pont G, Degroote F, Picard G. Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes[J]. J Mol Biol, 1987, 195(2):447-451.
pmid: 3116263 |
| [22] |
Regev A, Cohen S, Cohen E, et al. Telomeric repeats on small polydisperse circular DNA(spcDNA)and genomic instability[J]. Oncogene, 1998, 17(26):3455-3461.
pmid: 10030669 |
| [23] |
Cohen S, Agmon N, Sobol O, et al. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells[J]. Mob DNA, 2010, 1(1):11.
doi: 10.1186/1759-8753-1-11 URL |
| [24] |
Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops[J]. Mol Cell Biol, 2004, 24(22):9948-9957.
doi: 10.1128/MCB.24.22.9948-9957.2004 URL |
| [25] |
Tomaska L, Nosek J, Kramara J, et al. Telomeric circles:universal players in telomere maintenance?[J]. Nat Struct Mol Biol, 2009, 16(10):1010-1015.
doi: 10.1038/nsmb.1660 URL |
| [26] |
Kunisada T, Yamagishi H. Sequence repetition and genomic distribution of small polydisperse circular DNA purified from HeLa cells[J]. Gene, 1984, 31(1/2/3):213-223.
doi: 10.1016/0378-1119(84)90212-9 URL |
| [27] |
Stanfield SW, Helinski DR. Cloning and characterization of small circular DNA from Chinese hamster ovary cells[J]. Mol Cell Biol, 1984, 4(1):173-180.
doi: 10.1128/mcb.4.1.173-180.1984 pmid: 6700583 |
| [28] |
Nielsen JL, Walsh JT, Degen DR, et al. Evidence of gene amplification in the form of double minute chromosomes is frequently observed in lung cancer[J]. Cancer Genet Cytogenet, 1993, 65(2):120-124.
doi: 10.1016/0165-4608(93)90219-C URL |
| [29] |
Fletcher JA, Gebhardt MC, Kozakewich HP. Cytogenetic aberrations in osteosarcomas. Nonrandom deletions, rings, and double-minute chromosomes[J]. Cancer Genet Cytogenet, 1994, 77(1):81-88.
doi: 10.1016/0165-4608(94)90154-6 URL |
| [30] |
Vogt N, Lefèvre SH, Apiou F, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas[J]. PNAS, 2004, 101(31):11368-11373.
doi: 10.1073/pnas.0402979101 URL |
| [31] |
Gibaud A, Vogt N, Hadj-Hamou NS, et al. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci[J]. Hum Mol Genet, 2010, 19(7):1276-1285.
doi: 10.1093/hmg/ddq004 pmid: 20056677 |
| [32] |
Reader JC, Zhao XF, Butler MS, et al. REL-positive double minute chromosomes in follicular lymphoma[J]. Leukemia, 2006, 20(9):1624-1626.
pmid: 16775615 |
| [33] |
Villa O, Salido M, Pérez-Vila ME, et al. Blast cells with nuclear extrusions in the form of micronuclei are associated with MYC amplification in acute myeloid leukemia[J]. Cancer Genet Cytogenet, 2008, 185(1):32-36.
doi: 10.1016/j.cancergencyto.2008.04.014 URL |
| [34] |
Benner SE, Wahl GM, von Hoff DD. Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines[J]. Anti Cancer Drugs, 1991, 2(1):11-25.
doi: 10.1097/00001813-199102000-00002 URL |
| [35] |
Hahn PJ. Molecular biology of double-minute chromosomes[J]. Bioessays, 1993, 15(7):477-484.
pmid: 7691058 |
| [36] |
Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia—a review[J]. Clin Transl Oncol, 2005, 7(11):477-485.
pmid: 16373058 |
| [37] |
Shibata Y, Kumar P, Layer R, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues[J]. Science, 2012, 336(6077):82-86.
doi: 10.1126/science.1213307 URL |
| [38] |
Møller HD, Mohiyuddin M, Prada-Luengo I, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue[J]. Nat Commun, 2018, 9(1):1069.
doi: 10.1038/s41467-018-03369-8 URL |
| [39] |
Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity[J]. Nature, 2017, 543(7643):122-125.
doi: 10.1038/nature21356 URL |
| [40] |
Verhaak RGW, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution[J]. Nat Rev Cancer, 2019, 19(5):283-288.
doi: 10.1038/s41568-019-0128-6 pmid: 30872802 |
| [41] |
Wu S, Turner KM, Nguyen N, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression[J]. Nature, 2019, 575(7784):699-703.
doi: 10.1038/s41586-019-1763-5 URL |
| [42] |
Koche RP, Rodriguez-Fos E, Helmsauer K, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma[J]. Nat Genet, 2020, 52(1):29-34.
doi: 10.1038/s41588-019-0547-z URL |
| [43] | Kinoshita Y, Ohnishi N, Yamada Y, et al. Extrachromosomal circular DNA from nuclear fraction of higher plants[J]. Plant and Cell Physiology, 1985, 26(7):1401-1409. |
| [44] | Kunisada T, Yamagishi H, Kinoshita I, et al. Amplification of extrachromosomal circular DNA in intact bean leaves treated with benzyladenine[J]. Plant Cell Physiol, 1986, 27(2):355-361. |
| [45] |
Carroll SM, Gaudray P, De Rose ML, et al. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency:functional evidence for a mammalian replication origin[J]. Mol Cell Biol, 1987, 7(5):1740-1750.
doi: 10.1128/mcb.7.5.1740-1750.1987 pmid: 2885742 |
| [46] |
Carroll SM, DeRose ML, Gaudray P, et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion[J]. Mol Cell Biol, 1988, 8(4):1525-1533.
doi: 10.1128/mcb.8.4.1525-1533.1988 pmid: 2898098 |
| [47] |
Ruiz JC, Choi KH, von Hoff DD, et al. Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line[J]. Mol Cell Biol, 1989, 9(1):109-115.
doi: 10.1128/mcb.9.1.109-115.1989 pmid: 2648129 |
| [48] | Daniel D Von Hoff. New mechanisms of gene amplification in drug resistance[M]// Ozols RF. Molecular and clinical advances in anticancer drug resistance. New York:Springer Science & Business Media, 2012:1-8. |
| [49] |
Schoenlein PV, Barrett JT, Kulharya A, et al. Radiation therapy depletes extrachromosomally amplified drug resistance genes and oncogenes from tumor cells via micronuclear capture of episomes and double minute chromosomes[J]. Int J Radiat Oncol Biol Phys, 2003, 55(4):1051-1065.
doi: 10.1016/S0360-3016(02)04473-5 URL |
| [50] |
Storlazzi CT, Fioretos T, Surace C, et al. MYC-containing double minutes in hematologic malignancies:evidence in favor of the episome model and exclusion of MYC as the target gene[J]. Hum Mol Genet, 2006, 15(6):933-942.
pmid: 16452126 |
| [51] |
Shimizu N. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements[J]. Mutagenesis, 2011, 26(1):119-123.
doi: 10.1093/mutage/geq053 URL |
| [52] |
Chiu RWK, Dutta A, Hensson AG, et al. What is extrachromosomal circular DNA and what does it do?[J]. Clin Chem, 2020, 66(6):754-759.
doi: 10.1093/clinchem/hvaa096 URL |
| [53] |
Hull RM, King M, Pizza G, et al. Transcription-induced formation of extrachromosomal DNA during yeast ageing[J]. PLoS Biol, 2019, 17(12):e3000471.
doi: 10.1371/journal.pbio.3000471 URL |
| [54] |
Dillon LW, Kumar P, Shibata Y, et al. Production of extrachromosomal MicroDNAs is linked to mismatch repair pathways and transcriptional activity[J]. Cell Rep, 2015, 11(11):1749-1759.
doi: 10.1016/j.celrep.2015.05.020 pmid: 26051933 |
| [55] |
Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats[J]. Mol Cell, 2019, 73(3):398-411.
doi: S1097-2765(19)30044-9 pmid: 30735654 |
| [56] |
Brewer BJ, Payen C, Di Rienzi SC, et al. Origin-dependent inverted-repeat amplification:tests of a model for inverted DNA amplification[J]. PLoS Genet, 2015, 11(12):e1005699.
doi: 10.1371/journal.pgen.1005699 URL |
| [57] |
Meng X, Qi X, Guo H, et al. Novel role for non-homologous end joining in the formation of double minutes in methotrexate-resistant colon cancer cells[J]. J Med Genet, 2015, 52(2):135-144.
doi: 10.1136/jmedgenet-2014-102703 URL |
| [58] |
Cai M, Zhang H, Hou L, et al. Inhibiting homologous recombination decreases extrachromosomal amplification but has no effect on intrachromosomal amplification in methotrexate-resistant colon cancer cells[J]. Int J Cancer, 2019, 144(5):1037-1048.
doi: 10.1002/ijc.v144.5 URL |
| [59] |
Nelson JO, Watase GJ, Warsinger-Pepe N, et al. Mechanisms of rDNA copy number maintenance[J]. Trends Genet, 2019, 35(10):734-742.
doi: 10.1016/j.tig.2019.07.006 URL |
| [60] |
Yerlici VT, Lu MW, Hoge CR, et al. Programmed genome rearrangements in Oxytricha produce transcriptionally active extrachromosomal circular DNA[J]. Nucleic Acids Res, 2019, 47(18):9741-9760.
doi: 10.1093/nar/gkz725 pmid: 31504770 |
| [61] |
Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase:keeping linear chromosomes via telomeric circles[J]. FEBS Lett, 2004, 567(1):142-146.
doi: 10.1016/j.febslet.2004.04.058 URL |
| [62] |
Park PU, Defossez PA, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae[J]. Mol Cell Biol, 1999, 19(5):3848-3856.
doi: 10.1128/MCB.19.5.3848 pmid: 10207108 |
| [63] |
Gresham D, Usaite R, Germann SM, et al. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus[J]. PNAS, 2010, 107(43):18551-18556.
doi: 10.1073/pnas.1014023107 pmid: 20937885 |
| [64] |
Paulsen T, Kumar P, Koseoglu MM, et al. Discoveries of extrachromosomal circles of DNA in normal and tumor cells[J]. Trends Genet, 2018, 34(4):270-278.
doi: 10.1016/j.tig.2017.12.010 URL |
| [65] |
Gu X, Yu J, Chai P, et al. Novel insights into extrachromosomal DNA:redefining the onco-drivers of tumor progression[J]. J Exp Clin Cancer Res, 2020, 39(1):215.
doi: 10.1186/s13046-020-01726-4 URL |
| [66] |
Yan YL, Guo GJ, Huang JZ, et al. Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance[J]. J Hematol Oncol, 2020, 13(1):124.
doi: 10.1186/s13045-020-00960-9 URL |
| [67] |
Liao Z, Jiang W, Ye L, et al. Classification of extrachromosomal circular DNA with a focus on the role of extrachromosomal DNA(ecDNA)in tumor heterogeneity and progression[J]. Biochim Biophys Acta Rev Cancer, 2020, 1874(1):188392.
doi: 10.1016/j.bbcan.2020.188392 URL |
| [68] | Mansisidor A, Molinar T Jr, Srivastava P Jr, et al. Genomic copy-number loss is rescued by self-limiting production of DNA circles[J]. Mol Cell, 2018, 72(3):583-593. e4. |
| [69] | Wei JX, Wu CC, Meng HB, et al. The biogenesis and roles of extrachromosomal oncogene involved in carcinogenesis and evolution[J]. Am J Cancer Res, 2020, 10(11):3532-3550. |
| [70] |
Ain Q, Schmeer C, Wengerodt D, et al. Extrachromosomal circular DNA:current knowledge and implications for CNS aging and neurodegeneration[J]. International Journal of Molecular Sciences, 2020, 21(7):2477.
doi: 10.3390/ijms21072477 URL |
| [71] | Qiu H, Shao ZY, Wen X, et al. New insights of extrachromosomal DNA in tumorigenesis and therapeutic resistance of cancer[J]. Am J Cancer Res, 2020, 10(12):4056-4065. |
| [72] |
Mazzucco G, Huda A, Galli M, et al. Telomere damage induces internal loops that generate telomeric circles[J]. Nat Commun, 2020, 11(1):5297.
doi: 10.1038/s41467-020-19139-4 URL |
| [73] |
Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer[J]. Cancer Lett, 2003, 194(2):155-162.
pmid: 12757973 |
| [74] |
Sinclair DA, Guarente L. Extrachromosomal rDNA circles——a cause of aging in yeast[J]. Cell, 1997, 91(7):1033-1042.
pmid: 9428525 |
| [75] |
Hull RM, Houseley J. The adaptive potential of circular DNA accumulation in ageing cells[J]. Curr Genet, 2020, 66(5):889-894.
doi: 10.1007/s00294-020-01069-9 URL |
| [76] |
Møller HD, Parsons L, Jørgensen TS, et al. Extrachromosomal circular DNA is common in yeast[J]. PNAS, 2015, 112(24):E3114-E3122.
doi: 10.1073/pnas.1508825112 URL |
| [77] |
Payen C, Sunshine AB, Ong GT, et al. High-throughput identification of adaptive mutations in experimentally evolved yeast populations[J]. PLoS Genet, 2016, 12(10):e1006339.
doi: 10.1371/journal.pgen.1006339 URL |
| [78] |
Nathanson DA, Gini B, Mottahedeh J, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA[J]. Science, 2014, 343(6166):72-76.
doi: 10.1126/science.1241328 pmid: 24310612 |
| [79] |
Nikolaev S, Santoni F, Garieri M, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma[J]. Nat Commun, 2014, 5:5690.
doi: 10.1038/ncomms6690 pmid: 25471132 |
| [80] |
DeCarvalho AC, Kim H, Poisson LM, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma[J]. Nat Genet, 2018, 50(5):708-717.
doi: 10.1038/s41588-018-0105-0 URL |
| [81] |
Bailey C, Shoura MJ, Mischel PS, et al. Extrachromosomal DNA-relieving heredity constraints, accelerating tumour evolution[J]. Ann Oncol, 2020, 31(7):884-893.
doi: S0923-7534(20)36392-4 pmid: 32275948 |
| [82] | Morton AR, Dogan-Artun N, Faber ZJ, et al. Functional enhancers shape extrachromosomal oncogene amplifications[J]. Cell, 2019, 179(6):1330-1341. e13. |
| [83] |
Ott CJ. Circles with a point:new insights into oncogenic extrachromosomal DNA[J]. Cancer Cell, 2020, 37(2):145-146.
doi: 10.1016/j.ccell.2020.01.008 URL |
| [84] |
Sin STK, Jiang P, Deng J, et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma[J]. PNAS, 2020, 117(3):1658-1665.
doi: 10.1073/pnas.1914949117 URL |
| [85] |
Kumar P, Dillon LW, Shibata Y, et al. Normal and cancerous tissues release extrachromosomal circular DNA(eccDNA)into the circulation[J]. Mol Cancer Res, 2017, 15(9):1197-1205.
doi: 10.1158/1541-7786.MCR-17-0095 URL |
| [86] |
Kim H, Nguyen NP, Turner K, et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers[J]. Nat Genet, 2020, 52(9):891-897.
doi: 10.1038/s41588-020-0678-2 URL |
| [87] | Tandon I, Pal R, Pal JK, et al. Extrachromosomal circular DNAs:an extra piece of evidence to depict tumor heterogeneity[J]. Future Sci OA, 2019, 5(6):FSO390. |
| [88] |
Cohen S, Regev A, Lavi S. Small polydispersed circular DNA(spcDNA)in human cells:association with genomic instability[J]. Oncogene, 1997, 14(8):977-985.
pmid: 9050997 |
| [89] |
Cohen Z, Bacharach E, Lavi S. Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway[J]. Oncogene, 2006, 25(33):4515-4524.
pmid: 16547499 |
| [90] |
Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes:possible involvement in the plasticity of tandem repeats[J]. Cytogenet Genome Res, 2009, 124(3/4):327-338.
doi: 10.1159/000218136 URL |
| [91] |
Hollis M, Hindley J. Human Sau3A repeated DNA is enriched in small polydisperse circular DNA from normal lymphocytes[J]. Gene, 1986, 46(2/3):153-160.
doi: 10.1016/0378-1119(86)90399-9 URL |
| [92] |
Shoura MJ, Gabdank I, Hansen L, et al. Intricate and cell Type-Specific populations of endogenous circular dna(eccDNA)in Caenorhabditis elegans and Homo sapiens[J]. G3 Genes|Genomes|Genetics, 2017, 7(10):3295-3303.
doi: 10.1534/g3.117.300141 URL |
| [93] |
Lanciano S, Carpentier MC, Llauro C, et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants[J]. PLoS Genet, 2017, 13(2):e1006630.
doi: 10.1371/journal.pgen.1006630 URL |
| [94] |
Mehta D, Cornet L, Hirsch-Hoffmann M, et al. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq[J]. Nat Protoc, 2020, 15(5):1673-1689.
doi: 10.1038/s41596-020-0301-0 URL |
| [95] | Kumar P, Kiran S, Saha S, et al. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines[J]. Sci Adv, 2020, 6(20):eaba2489. |
| [96] |
Deshpande V, Luebeck J, Nguyen ND, et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect[J]. Nat Commun, 2019, 10(1):392.
doi: 10.1038/s41467-018-08200-y pmid: 30674876 |
| [97] |
Luebeck J, Coruh C, Dehkordi SR, et al. AmpliconReconstructor integrates NGS and optical mapping to resolve the complex structures of focal amplifications[J]. Nat Commun, 2020, 11(1):4374.
doi: 10.1038/s41467-020-18099-z pmid: 32873787 |
| [98] |
Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs[J]. Genome Res, 2016, 26(9):1277-1287.
doi: 10.1101/gr.202895.115 URL |
| [99] |
Prada-Luengo I, Krogh A, Maretty L, et al. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads[J]. BMC Bioinform, 2019, 20(1):1-9.
doi: 10.1186/s12859-018-2565-8 URL |
| [100] | Mann L, Seibt KM, Weber B, et al. Biopolis Dresden PhD Symposium 2020[C]. Dresden, 2020. |
| [101] |
Yu J, Xiang X, Huang J, et al. Haplotyping by CRISPR-mediated DNA circularization(CRISPR-hapC)broadens allele-specific gene editing[J]. Nucleic Acids Res, 2020, 48(5):e25.
doi: 10.1093/nar/gkz1233 URL |
| [102] |
Iparraguirre L, Prada-Luengo I, Regenberg B, et al. To be or not to be:circular RNAs or mRNAs from circular DNAs?[J]. Front Genet, 2019, 10:940.
doi: 10.3389/fgene.2019.00940 pmid: 31681407 |
| [103] |
Gaines TA, Patterson EL, Neve P. Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance[J]. New Phytol, 2019, 223(4):1770-1775.
doi: 10.1111/nph.15858 pmid: 31002387 |
| [1] | 肖亮, 吴正丹, 陆柳英, 施平丽, 尚小红, 曹升, 曾文丹, 严华兵. 木薯重要性状基因的研究进展[J]. 生物技术通报, 2023, 39(6): 31-48. |
| [2] | 孙海航, 官会林, 王旭, 王童, 李泓霖, 彭文洁, 刘柏桢, 樊芳玲. 生物炭对三七连作土壤性质及真菌群落的影响[J]. 生物技术通报, 2023, 39(2): 221-231. |
| [3] | 王子夜, 王志刚, 阎爱华. 不同树龄桑根际土壤原生生物群落组成多样性[J]. 生物技术通报, 2022, 38(8): 206-215. |
| [4] | 陈天赐, 武少兰, 杨国辉, 江丹霞, 江玉姬, 陈炳智. 无柄灵芝醇提物对小鼠睡眠及肠道菌群的影响[J]. 生物技术通报, 2022, 38(8): 225-232. |
| [5] | 钟辉, 刘亚军, 王滨花, 和梦洁, 吴兰. 分析方法对细菌群落16S rRNA基因扩增测序分析结果的影响[J]. 生物技术通报, 2022, 38(6): 81-92. |
| [6] | 杨亚杰, 李昱樱, 申状状, 陈天, 荣二花, 吴玉香. 草棉同源多倍体后代筛选及性状鉴定[J]. 生物技术通报, 2022, 38(5): 64-73. |
| [7] | 赵林艳, 官会林, 向萍, 李泽诚, 柏雨龙, 宋洪川, 孙世中, 徐武美. 白及根腐病植株根际土壤微生物群落组成特征分析[J]. 生物技术通报, 2022, 38(2): 67-74. |
| [8] | 陈宇捷, 郑华宝, 周昕彦. 改良高通量测序技术揭示除藻剂对藻类群落的影响[J]. 生物技术通报, 2022, 38(11): 70-79. |
| [9] | 孔德真, 聂迎彬, 徐红军, 崔凤娟, 穆培源, 田笑明. 三系杂交小麦混播制种对杂交种产量、纯度及F1产量优势的影响[J]. 生物技术通报, 2022, 38(10): 132-139. |
| [10] | 毛婷, 牛永艳, 郑群, 杨涛, 穆永松, 祝英, 季彬, 王治业. 菌剂对苜蓿青贮发酵品质及微生物群落的影响[J]. 生物技术通报, 2021, 37(9): 86-94. |
| [11] | 唐蝶, 周倩. 植物基因组组装技术研究进展[J]. 生物技术通报, 2021, 37(6): 1-12. |
| [12] | 吕燕, 刘建利, 李靖宇, 候琳琳, 孙敏, 苟琪. 不同品种和产区宁夏枸杞根系AMF多样性[J]. 生物技术通报, 2021, 37(6): 36-48. |
| [13] | 朱斌, 甘晨晨, 王洪程. 球花石斛(Dendrobium thyrsiflorum)叶绿体基因组特征及亲缘关系解析[J]. 生物技术通报, 2021, 37(5): 38-47. |
| [14] | 孙平勇, 张武汉, 舒服, 何强, 张莉, 彭志荣, 邓华凤. 香稻品种OsBADH2突变位点分析及其功能标记的开发[J]. 生物技术通报, 2021, 37(4): 1-7. |
| [15] | 张秫华, 方千, 贾红梅, 韩桂琪, 严铸云, 何冬梅. 川芎非根际、根际及根茎内生真菌群落差异分析[J]. 生物技术通报, 2021, 37(4): 56-69. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||