








生物技术通报 ›› 2022, Vol. 38 ›› Issue (8): 198-205.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1482
收稿日期:2021-11-29
出版日期:2022-08-26
发布日期:2022-09-14
作者简介:唐光甫,男,硕士研究生,研究方向:药用微生物;E-mail: 基金资助:
TANG Guang-fu( ), GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong(
), GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong( )
)
Received:2021-11-29
Published:2022-08-26
Online:2022-09-14
摘要:
为研究红曲霉pyrG基因对其次生代谢的影响,通过农杆菌介导遗传转化获得了转Cas 9基因的红曲霉底盘菌株,然后设计并体外转录合成pyrG基因的定向编辑sgRNA,对底盘菌株的原生质体遗传转化,经5-FOA抗性筛选、PCR扩增、T7EI酶切和测序验证,获得pyrG基因定向编辑的红曲霉4个菌株,其中2株单碱基缺失,2株2碱基缺失。比较分析表明,野生型红曲霉在添加尿嘧啶核苷的培养基中会极显著增加洛伐他汀含量,并极显著降低桔霉素含量,而经CRISPR/Cas 9失活pyrG的变异菌株与野生型相比,洛伐他汀和桔霉素含量明显增加。表明尿嘧啶核苷合成关键酶基因pyrG和外源尿嘧啶核苷对红曲霉的次生代谢有协同调控作用,为红曲霉功能基因研究和次生代谢调控提供参考。
唐光甫, 桂艳玲, 满海乔, 赵杰宏. 利用CRISPR/Cas 9编辑红曲霉pyrG基因对其次生代谢的影响[J]. 生物技术通报, 2022, 38(8): 198-205.
TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism[J]. Biotechnology Bulletin, 2022, 38(8): 198-205.
| 引物名称 Primer name | 引物或sgRNA序列 Primer or sgRNA sequence(5'-3') | 退火温度 Annea-ling temperature/℃ | 靶标序列长度 Target sequence length/bp |
|---|---|---|---|
| Cas9-F | GTTTAAGGTCCTGGGCAACA | 55 | 312 |
| Cas9-R | ATCGTGGGGTACTTCTCGTG | ||
| PyrGgr-F | GTTGAAACGAACCCCAGCAC | 57 | 380 |
| pyrGgr-R | CCACATCGACATCCTCTCCG | ||
| sgRNA | TTAATACGACTCACTATAGGGggcttgaagttcctgcgttgGTTTTAGAGCTAGAAATA | / | / |
表1 PCR检测引物和sgRNA序列
Table 1 Primers and sgRNA sequences for PCR detection
| 引物名称 Primer name | 引物或sgRNA序列 Primer or sgRNA sequence(5'-3') | 退火温度 Annea-ling temperature/℃ | 靶标序列长度 Target sequence length/bp |
|---|---|---|---|
| Cas9-F | GTTTAAGGTCCTGGGCAACA | 55 | 312 |
| Cas9-R | ATCGTGGGGTACTTCTCGTG | ||
| PyrGgr-F | GTTGAAACGAACCCCAGCAC | 57 | 380 |
| pyrGgr-R | CCACATCGACATCCTCTCCG | ||
| sgRNA | TTAATACGACTCACTATAGGGggcttgaagttcctgcgttgGTTTTAGAGCTAGAAATA | / | / |

图3 农杆菌介导Cas 9转化红曲霉 A:农杆菌PCR检测;B:农杆菌和红曲霉共培养;C:红曲霉Cas 9底盘菌株PCR检测;1-8:不同菌株;M:DL1000 DNA Marker
Fig. 3 Transformation of Cas 9 into monascus mediated by A. tumefaciens A:PCR detection of A. tumefaciens. B:Co-culture of A. tumefaciens and monascus. C:PCR detection of the Cas 9 of monascus chassis strain. 1-8:Different strains. M:DL1000 DNA marker
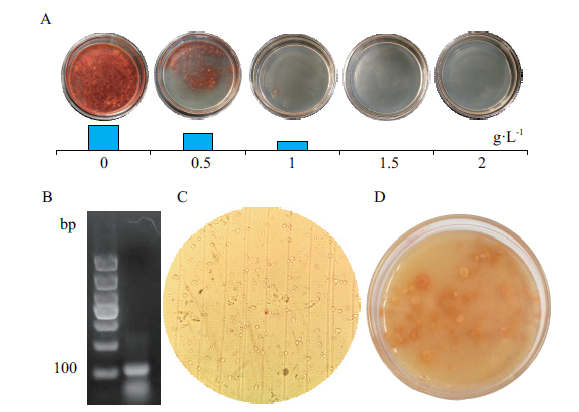
图4 体外sgRNA转化红曲霉原生质体 A:5-FOA对红曲霉的抑制作用;B:sgRNA电泳检测;C:制备的原生质体;D:抗性红曲霉筛选
Fig. 4 Transformation of monascus protoplasts by sgRNA in vitro A:Inhibition of 5-FOA on monascus. B:Electrophoresis of the sgRNA. C:Prepared protoplasts. D:Screening resistant monascus
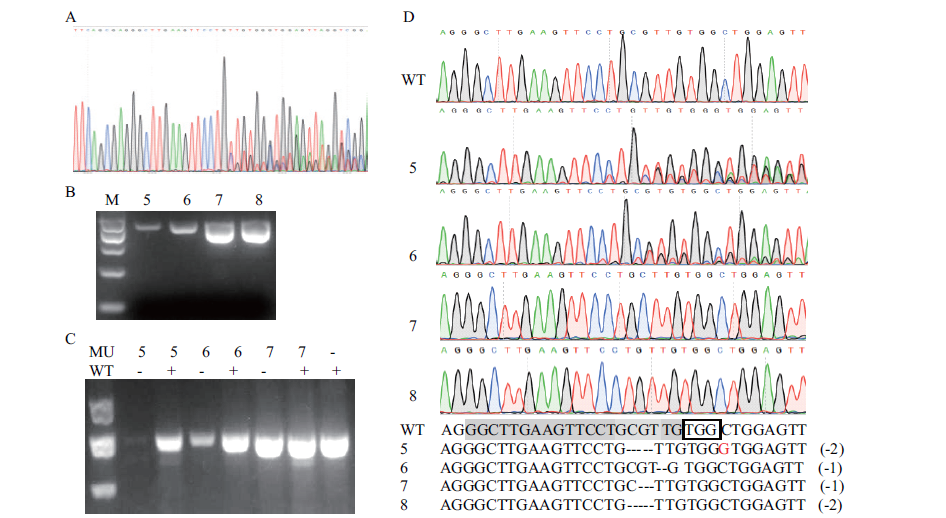
图5 红曲霉突变株pyrG基因的变异分析 A:局部套峰序列;B:部分菌株pyrG基因的PCR检测;C:T7EI酶切PCR产物;D:部分菌株pyrG基因的变异位点。MU:突变株;WT:野生型;“+”:有WT DNA;“-”:无WT DNA;方框:PAM;灰底色:sgRNA靶标序列
Fig. 5 Variation analysis of pyrG gene of monascus mutant A:Part nested peak sequence. B:PCR detection of pyrG gene in some strains. C:PCR product cutted by T7EI. D:Mutation loci of pyrG gene in some strains. MU:Mutant. WT:Wild-type. “+”:With DNA. “-”:Without DNA. Black box:Refers to the PAM site. Grey background:Refers to the sgRNA target sequence
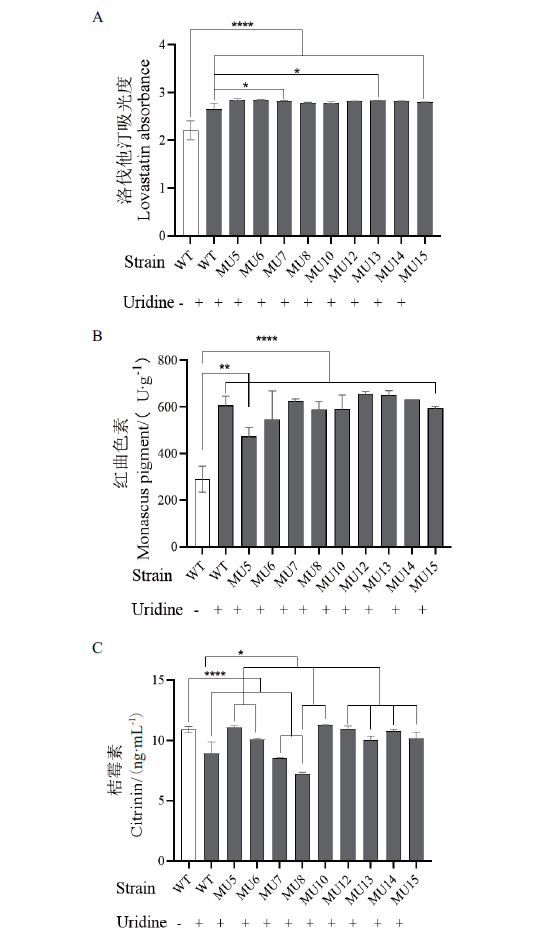
图7 培养基中添加尿嘧啶核苷对不同菌株化学成分含量的影响 A:洛伐他汀吸光度;B:红曲色素含量;C:桔霉素含量;“-”:表示未添加;“+”:表示添加;WT:野生型;MU:变异菌
Fig. 7 Effect of uracil riboside on the chemical composition content of different strains in medium A:Lovastatin absorbance. B:Monascus pigment content. C:Citrinin content. “-”:Not added. “+”:Added. WT:Wild type strain. MU:Mutant strain
| [1] | 傅金泉. 中国红曲及其实用技术[M]. 北京: 中国轻工业出版社, 1997. |
| Fu JQ. Chinese red yeast rice and its practical technology[M]. Beijing: China Light Industry Press, 1997. | |
| [2] | 李利, 陈莎, 陈福生, 等. 红曲菌次生代谢产物生物合成途径及相关基因的研究进展[J]. 微生物学通报, 2013, 40(2):294-303. |
| Li L, Chen S, Chen FS, et al. Review on biosynthetic pathway of secondary metabolites and the related genes in Monascus spp[J]. Microbiol China, 2013, 40(2):294-303. | |
| [3] |
Blanc PJ, Laussac JP, Bars JL, et al. Characterization of monascidin A from Monascus as citrinin[J]. Int J Food Microbiol, 1995, 27(2/3):201-213.
doi: 10.1016/0168-1605(94)00167-5 URL |
| [4] | 李贞景, 薛意斌, 刘妍, 等. 红曲菌中桔霉素的控制策略及研究进展[J]. 食品科学, 2018, 39(17):263-268. |
| Li ZJ, Xue YB, Liu Y, et al. Recent progress on control strategies against citrinin in Monascus spp[J]. Food Sci, 2018, 39(17):263-268. | |
| [5] |
Shimizu T, Kinoshita H, Nihira T. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus[J]. Appl Environ Microbiol, 2007, 73(16):5097-5103.
doi: 10.1128/AEM.01979-06 URL |
| [6] |
Li YP, Pan YF, Zou LH, et al. Lower citrinin production by gene disruption of ctnB involved in citrinin biosynthesis in Monascus aurantiacus Li AS3. 4384[J]. J Agric Food Chem, 2013, 61(30):7397-7402.
doi: 10.1021/jf400879s URL |
| [7] |
Ning ZQ, Cui H, Xu Y, et al. Deleting the citrinin biosynthesis-related gene, ctnE, to greatly reduce citrinin production in Monascus aurantiacus Li AS3. 4384[J]. Int J Food Microbiol, 2017, 241:325-330.
doi: 10.1016/j.ijfoodmicro.2016.11.004 URL |
| [8] | Li YP, Wang N, Jiao XX, et al. The ctnF gene is involved in citrinin and pigment synthesis in Monascus aurantiacus[J]. J Basic Microbiol, 2020, 60(10):873-881. |
| [9] |
Li YP, Tang X, Wu W, et al. The ctnG gene encodes carbonic anhydrase involved in mycotoxin citrinin biosynthesis from Monascus aurantiacus[J]. Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2015, 32(4):577-583.
doi: 10.1080/19440049.2014.990993 URL |
| [10] | 吴伟. 橙色红曲菌ctnG基因和ctnH基因缺失菌株的构建及其功能分析[D]. 南昌: 南昌大学, 2010. |
| Wu W. Construction and functional analysis of the ctnG and ctnH gene disruption mutants in Monascus aurantiacus AS3. 4384[D]. Nanchang: Nanchang University, 2010. | |
| [11] |
Balakrishnan B, Chandran R, Park SH, et al. Delineating citrinin biosynthesis:Ctn-ORF3 dioxygenase-mediated multi-step methyl oxidation precedes a reduction-mediated pyran ring cyclization[J]. Bioorg Med Chem Lett, 2016, 26(2):392-396.
doi: S0960-894X(15)30314-0 pmid: 26707397 |
| [12] | 崔华. 橙色红曲菌桔霉素合成相关基因-orf3和ctnE缺失菌株的构建及其相关分析[D]. 南昌: 南昌大学, 2012. |
| Cui H. Construction and correlation analysis of the Orf3and ctnE gene disruption mutants in Monascus aurantiacus AS3. 4384[D]. Nanchang: Nanchang University, 2012. | |
| [13] |
Liang B, Du XJ, Li P, et al. Orf6 gene encoded glyoxalase involved in mycotoxin citrinin biosynthesis in Monascus purpureus YY-1[J]. Appl Microbiol Biotechnol, 2017, 101(19):7281-7292.
doi: 10.1007/s00253-017-8462-7 pmid: 28831532 |
| [14] | 邹乐花, 李燕萍, 黄志兵, 等. 橙色红曲菌As3. 4384 orf7基因缺失株的构建及其功能分析[J]. 中国生物工程杂志, 2011, 31(7):79-84. |
| Zou LH, Li YP, Huang ZB, et al. Construction of the Monascus aurantiacus As3. 4384 orf7 gene deletion strains and functional analysis[J]. China Biotechnol, 2011, 31(7):79-84. | |
| [15] |
Shimizu T, Kinoshita H, Ishihara S, et al. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus[J]. Appl Environ Microbiol, 2005, 71(7):3453-3457.
doi: 10.1128/AEM.71.7.3453-3457.2005 URL |
| [16] |
Arbour CA, Imperiali B. Uridine natural products:challenging targets and inspiration for novel small molecule inhibitors[J]. Bioorg Med Chem, 2020, 28(18):115661.
doi: 10.1016/j.bmc.2020.115661 URL |
| [17] |
Zhang LH, Zheng XM, Cairns TC, et al. Disruption or reduced expression of the orotidine-5'-decarboxylase gene pyrG increases citric acid production:a new discovery during recyclable genome editing in Aspergillus niger[J]. Microb Cell Fact, 2020, 19(1):76.
doi: 10.1186/s12934-020-01334-z URL |
| [18] | 王汝毅. 根癌农杆菌介导红曲霉转化库的构建及转化子性质研究[D]. 武汉: 华中农业大学, 2005. |
| Wang RY. Construction of transformation library of Monascus ruber mediated by Agrobacterium tumefaciens and study on transformants’ characters[D]. Wuhan: Huazhong Agricultural University, 2005. | |
| [19] | 周礼红, 李国琴, 王正祥, 等. 红曲霉原生质体的制备、再生及其遗传转化系统[J]. 遗传, 2005, 27(3):423-428. |
| Zhou LH, Li GQ, Wang ZX, et al. Preparation and regeneration of protoplasts from Monascus purpureus and genetic transformation system[J]. Hered Beijing, 2005, 27(3):423-428. | |
| [20] | 李亮. 高产洛伐他汀红曲菌株的选育及发酵条件研究[D]. 福州: 福州大学, 2016. |
| Li L. Research of breeding and fermentation conditions of high-lovastatin Monascus strains[D]. Fuzhou: Fuzhou University, 2016. | |
| [21] |
Liu R, Chen L, Jiang YP, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discov, 2015, 1:15007.
doi: 10.1038/celldisc.2015.7 URL |
| [22] |
Shen B, Zhang J, Wu HY, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting[J]. Cell Res, 2013, 23(5):720-723.
doi: 10.1038/cr.2013.46 pmid: 23545779 |
| [23] |
Song RJ, Zhai Q, Sun L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi:progress and perspective[J]. Appl Microbiol Biotechnol, 2019, 103(17):6919-6932.
doi: 10.1007/s00253-019-10007-w URL |
| [24] |
O’Donovan GA, Neuhard J. Pyrimidine metabolism in microorganisms[J]. Bacteriol Rev, 1970, 34(3):278-343.
doi: 10.1128/br.34.3.278-343.1970 URL |
| [1] | 潘国强, 吴思源, 刘璐, 郭惠明, 程红梅, 苏晓峰. 大丽轮枝菌(Verticillim dahliae)突变体库的构建与分析[J]. 生物技术通报, 2023, 39(5): 112-119. |
| [2] | 石佳, 朱秀梅, 薛梦雨, 余超, 魏一鸣, 杨凤环, 陈华民. 基于水稻原生质体的染色质免疫共沉淀技术优化及应用[J]. 生物技术通报, 2022, 38(7): 62-69. |
| [3] | 梁玲, 黄钦耿, 翁雪清, 吴松刚, 黄建忠. 产L-谷氨酸工程菌株的诱变选育及其发酵效率[J]. 生物技术通报, 2020, 36(6): 143-149. |
| [4] | 潘旭耀, 魏韬, 谭柱豪, 林龙镇, 郭丽琼, 林俊芳. 芽孢杆菌种间原生质体融合选育高产Surfactin新菌株[J]. 生物技术通报, 2019, 35(8): 238-245. |
| [5] | 赵小强, 陈志荣, 何芳, 沈楠, 高峰, 黄家风. 大丽轮枝菌原生质体的制备及再生[J]. 生物技术通报, 2018, 34(7): 166-173. |
| [6] | 孙晓瑞, 陈博文, 张小林, 孟俊龙. 秀珍菇单孢子菌株原生质体制备条件的优化[J]. 生物技术通报, 2018, 34(4): 70-76. |
| [7] | 王伟伟, 肖燕, 张艺夕, 刘晶, 唐唯, 李灿辉. 马铃薯晚疫病菌原生质体制备及再生体系的研究[J]. 生物技术通报, 2018, 34(4): 77-82. |
| [8] | 张晓慧,韩榕. 两种瞬时表达体系研究拟南芥Profilin-1的亚细胞定位[J]. 生物技术通报, 2017, 33(5): 57-62. |
| [9] | 陈楠, 于飞, 何艳柳, 卜宁. 一株水稻促生长内生真菌的绿色荧光蛋白基因标记与示踪[J]. 生物技术通报, 2017, 33(3): 100-105. |
| [10] | 林艳梅, 李瑞杰, 张慧杰, 秦秀林, 冯家勋. 纤维素酶高产菌株Trichoderma atroviride HP35-3原生质体制备及转化[J]. 生物技术通报, 2016, 32(9): 225-231. |
| [11] | 周明明, 李晓雁, 任梦楠, 程珂萌, 黄海东. 韦兰胶合成菌的原生质体制备与再生研究[J]. 生物技术通报, 2016, 32(7): 126-130. |
| [12] | 席海秀,艾可筠,佟少明. 普通小麦幼苗叶肉细胞原生质体分离方法的优化[J]. 生物技术通报, 2016, 32(4): 68-73. |
| [13] | 苟敏, 杨白雪, 汤岳琴, 木田建次. 利用原生质体融合技术构建耐酸絮凝性产乙醇酿酒酵母[J]. 生物技术通报, 2016, 32(11): 115-123. |
| [14] | 呼吉雅, 卢萍, 牛艳芳. 小花棘豆Embellisia内生真菌原生质体的制备与再生研究[J]. 生物技术通报, 2015, 31(5): 134-139. |
| [15] | 张晓立, 郑小梅, 满云, 罗虎, 于建东, 郑平, 刘浩, 孙际宾. 黑曲霉柠檬酸工业菌株原生质体制备与转化[J]. 生物技术通报, 2015, 31(3): 171-177. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||