








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 288-296.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0917
姚近东1,2,3( ), 汤华妹1,2,3, 杨文霄1,2,3, 张丽珊1,2,3, 林向民1,2,3(
), 汤华妹1,2,3, 杨文霄1,2,3, 张丽珊1,2,3, 林向民1,2,3( )
)
收稿日期:2022-07-24
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
林向民,男,博士,教授,研究方向:微生物分子生态学;E-mail:xiangmin@fafu.edu.cn作者简介:姚近东,男,研究方向 :微生物分子生态学 ;E-mail :945403230@qq.com汤华妹同为本文第一作者
基金资助:
YAO Jin-dong1,2,3( ), TANG Hua-mei1,2,3, YANG Wen-xiao1,2,3, ZHANG Li-shan1,2,3, LIN Xiang-min1,2,3(
), TANG Hua-mei1,2,3, YANG Wen-xiao1,2,3, ZHANG Li-shan1,2,3, LIN Xiang-min1,2,3( )
)
Received:2022-07-24
Published:2023-04-26
Online:2023-05-16
摘要:
为深入了解喹诺酮类抗生素胁迫下细菌产生的耐药机制,本研究以水生病原菌嗜水气单胞菌为研究对象,利用定量蛋白质组学方法比较嗜水气单胞菌在有无恩诺沙星处理下的蛋白表达差异。质谱结果发现446个差异蛋白,其中233个蛋白表达上调,213个蛋白表达下调。生物信息学分析显示,嗜水气单胞菌可能通过DNA修复和硫代谢相关蛋白的上调表达,促进细菌存活。进一步通过生长曲线测定发现,参与硫代谢的半胱氨酸与恩诺沙星联用能更好地抑制细菌的生长。同时,利用qPCR技术验证硫代谢相关基因在mRNA水平的表达量,发现大部分相关基因在转录水平上的差异表达与蛋白水平一致。以上研究结果表明硫代谢对嗜水气单胞菌在ENR胁迫下起着重要作用。
姚近东, 汤华妹, 杨文霄, 张丽珊, 林向民. 恩诺沙星胁迫下嗜水气单胞菌的比较蛋白质组学研究[J]. 生物技术通报, 2023, 39(4): 288-296.
YAO Jin-dong, TANG Hua-mei, YANG Wen-xiao, ZHANG Li-shan, LIN Xiang-min. Comparative Proteomics Analysis of Aeromonas hydrophila Under Enrofloxacin Stress[J]. Biotechnology Bulletin, 2023, 39(4): 288-296.
| Gene | Oligonucleotide sequence(5'-3') | Purpose |
|---|---|---|
| cysA-F | ATCGGGTGGTGCTGATGAACG | RT-qPCR |
| cysA-R | TGGGTAGCAGGCGGGTGATG | RT-qPCR |
| cysN-F | GGAGTCCGACAGCCAGAAG | RT-qPCR |
| cysN-R | GGCGGTGGAGAAATAGCG | RT-qPCR |
| cysD-F | GCTGGACATCTGGCAATACA | RT-qPCR |
| cysD-R | GCTCCTGCTTCACTTCATCTT | RT-qPCR |
| cysI-F | TCCACGCCAACGATCTCAA | RT-qPCR |
| cysI-R | GCCACGGCTGGTAAACTCAT | RT-qPCR |
| cysH-F | CCCCATCAATCGGGCTCT | RT-qPCR |
| cysH-R | TCCATCGGCTCCACCTTG | RT-qPCR |
| cysG2-F | CGCCTTTGCCTCCAACACC | RT-qPCR |
| cysG2-R | CCTTCTTGCCGACCGACACC | RT-qPCR |
| cysC-F | TATCGTGCTCACCGCCTTTA | RT-qPCR |
| cysC-R | CCTGCCCTAGCCTTCTTGTAG | RT-qPCR |
| cysK-F | ATCGGCGCCAACCTGATCT | RT-qPCR |
| cysK-R | CATTGCCAAGGCCAACGAG | RT-qPCR |
表1 本研究所使用的引物对列表
Table 1 Primer pairs used in this study
| Gene | Oligonucleotide sequence(5'-3') | Purpose |
|---|---|---|
| cysA-F | ATCGGGTGGTGCTGATGAACG | RT-qPCR |
| cysA-R | TGGGTAGCAGGCGGGTGATG | RT-qPCR |
| cysN-F | GGAGTCCGACAGCCAGAAG | RT-qPCR |
| cysN-R | GGCGGTGGAGAAATAGCG | RT-qPCR |
| cysD-F | GCTGGACATCTGGCAATACA | RT-qPCR |
| cysD-R | GCTCCTGCTTCACTTCATCTT | RT-qPCR |
| cysI-F | TCCACGCCAACGATCTCAA | RT-qPCR |
| cysI-R | GCCACGGCTGGTAAACTCAT | RT-qPCR |
| cysH-F | CCCCATCAATCGGGCTCT | RT-qPCR |
| cysH-R | TCCATCGGCTCCACCTTG | RT-qPCR |
| cysG2-F | CGCCTTTGCCTCCAACACC | RT-qPCR |
| cysG2-R | CCTTCTTGCCGACCGACACC | RT-qPCR |
| cysC-F | TATCGTGCTCACCGCCTTTA | RT-qPCR |
| cysC-R | CCTGCCCTAGCCTTCTTGTAG | RT-qPCR |
| cysK-F | ATCGGCGCCAACCTGATCT | RT-qPCR |
| cysK-R | CATTGCCAAGGCCAACGAG | RT-qPCR |
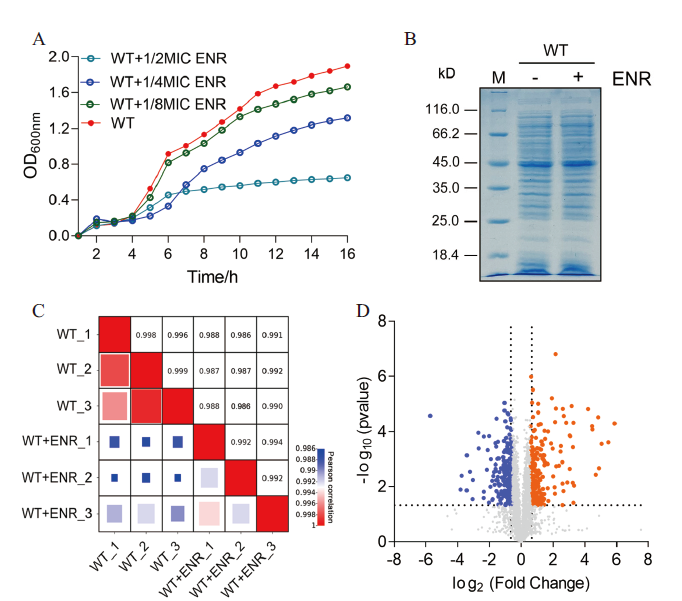
图1 定量蛋白质组学数据分析 A: 不同浓度恩诺沙星胁迫下嗜水气单胞菌的生长曲线;B: 嗜水气单胞菌在有无恩诺沙星处理下的SDS-PAGE图谱;C: 通过相关系数对各组样品的3次生物学重复的蛋白表达量进行相关性分析;D: 火山图显示蛋白表达差异的丰度。其中蓝色点表示差异表达下调的蛋白、橙色点表示差异表达上调的蛋白和灰色表达非差异表达的蛋白
Fig. 1 Analysis of quantitative proteomics data A: Growth curves of A. hydrophila under different concentrations of enrofloxacin stress. B: SDS-PAGE profiles of A. hydrophila with or without enrofloxacin treatment. C: Correlation analysis of protein expression of three biological replicates in each group was carried out by correlation coefficient. D: Volcanic plots showing the abundance of differential protein expression. The blue dots represent the differentially downregulated proteins, the orange dots represent the differentially up-regulated proteins, and the gray dots represent the non-differentially expressed proteins

图2 ENR胁迫下差异表达蛋白的GO和KEGG富集分析 A:使用DAVID在线网站和R语言分析差异表达蛋白的生物过程;B:分子功能;C:KEGG代谢通路富集分析
Fig. 2 GO and KEGG enrichment analysis of differentially expressed proteins under ENR stress A: Using DAVID online website and R language to analyze the biological processes of differentially expressed proteins. B: Molecular function. C: Enrichment analysis of KEGG metabolic pathway

图3 在ENR胁迫下差异蛋白的蛋白-蛋白相互作用预测(PPI)网络 核心蛋白扩展的网络。不同颜色表示差异蛋白的变化倍数(Log2)
Fig. 3 Protein-protein interaction prediction(PPI)net-work of differential proteins under ENR stress The extended network of hub proteins. Different colors indicate the fold change of different proteins(Log2)
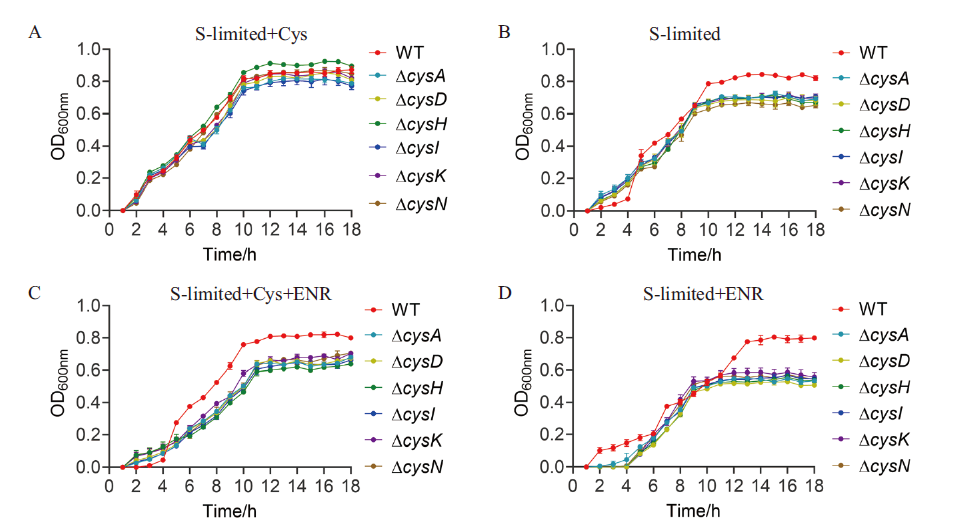
图4 WT和硫代谢相关基因缺失菌株在硫限制培养基中分别添加2 mmol/L半胱氨酸和ENR后的生长情况 A: 硫限制培养基中加入2 mmol/L半胱氨酸;B: 硫限制培养基; C: 硫限制培养基中加入2 mmol/L半胱氨酸和0.78 µg/mL ENR;D: 硫限制培养基中加入ENR
Fig. 4 Growths of WT and sulfur metabolism-related gene deletion strains supplemented with 2 mmol/L cysteine and ENR in sulfur limiting medium, respectively A: 2 mmol/L cysteine was added to the sulfur-limited medium. B: Sulfur-limited medium. C: Sulfur limited medium with 2 mmol/L cysteine and 0.78 µg/mL ENR. D: ENR was added to the sulfur-limited medium

图5 蛋白质组学数据的qPCR验证 采用qPCR方法验证嗜水气单胞菌在有无ENR胁迫下8个硫代谢相关基因在mRNA水平上的表达量
Fig. 5 qPCR validation of proteomic data qPCR was used to verify the mRNA expression levels of 8 sulfur metabolism related genes in A. hydrophila under ENR stress or not
| [1] |
Harikrishnan R, Balasundaram C. Modern trends in Aeromonas hydrophila disease management with fish[J]. Rev Fish Sci, 2005, 13(4): 281-320.
doi: 10.1080/10641260500320845 URL |
| [2] |
Adamski J, Koivuranta M, Leppänen E. Fatal case of myonecrosis and septicaemia caused by Aeromonas hydrophila in Finland[J]. Scand J Infect Dis, 2006, 38(11-12): 1117-1119.
pmid: 17148092 |
| [3] |
Dahanayake PS, Hossain S, Wickramanayake MVKS, et al. Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam(Ruditapes philippinarum)in Korea[J]. J Appl Microbiol, 2019, 127(3): 941-952.
doi: 10.1111/jam.14355 pmid: 31211903 |
| [4] |
Stratev D, Odeyemi OA. Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: a mini-review[J]. J Infect Public Health, 2016, 9(5): 535-544.
doi: 10.1016/j.jiph.2015.10.006 URL |
| [5] |
Paudel S, Peña-Bahamonde J, Shakiba S, et al. Prevention of infection caused by enteropathogenic E. coli O157: H7 in intestinal cells using enrofloxacin entrapped in polymer based nanocarriers[J]. J Hazard Mater, 2021, 414: 125454.
doi: 10.1016/j.jhazmat.2021.125454 URL |
| [6] |
Mitchell MA. Enrofloxacin[J]. J Exot Pet Med, 2006, 15(1): 66-69.
doi: 10.1053/j.jepm.2005.11.011 URL |
| [7] |
Sun J, Huang XY, Dong YP, et al. Transcriptome responses in enrofloxacin-resistant strains of Aeromonas hydrophila exposed to Forsythia suspensa[J]. Aquaculture, 2020, 517: 734809.
doi: 10.1016/j.aquaculture.2019.734809 URL |
| [8] |
Zhu FJ, Yang ZY, Zhang YL, et al. Transcriptome differences between enrofloxacin-resistant and enrofloxacin-susceptible strains of Aeromonas hydrophila[J]. PLoS One, 2017, 12(7): e0179549.
doi: 10.1371/journal.pone.0179549 URL |
| [9] |
Sha S, Dong ZX, Gao YS, et al. In-situ removal of residual antibiotics(enrofloxacin)in recirculating aquaculture system: effect of ultraviolet photolysis plus biodegradation using immobilized microbial granules[J]. J Clean Prod, 2022, 333: 130190.
doi: 10.1016/j.jclepro.2021.130190 URL |
| [10] |
Tanca A, Biosa G, Pagnozzi D, et al. Comparison of detergent-based sample preparation workflows for LTQ-Orbitrap analysis of the Escherichia coli proteome[J]. Proteomics, 2013, 13(17): 2597-2607.
doi: 10.1002/pmic.201200478 URL |
| [11] |
Wang GB, Wang YQ, Zhang LS, et al. Proteomics analysis reveals the effect of Aeromonas hydrophila sirtuin CobB on biological functions[J]. J Proteomics, 2020, 225: 103848.
doi: 10.1016/j.jprot.2020.103848 URL |
| [12] |
Huang DW, Sherman BT, Tan QN, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists[J]. Nucleic Acids Res, 2007, 35(Web Server issue): W169-W175.
doi: 10.1093/nar/gkm415 pmid: 17576678 |
| [13] |
Li Z, Zhang LS, Sun LN, et al. Proteomics analysis reveals the importance of transcriptional regulator slyA in regulation of several physiological functions in Aeromonas hydrophila[J]. J Proteomics, 2021, 244: 104275.
doi: 10.1016/j.jprot.2021.104275 URL |
| [14] | Yu J, Ramanathan S, Chen LC, et al. Comparative transcriptomic analysis reveals the molecular mechanisms related to oxytetracycline- resistance in strains of Aeromonas hydrophila[J]. Aquac Rep, 2021, 21: 100812. |
| [15] |
Cai QL, Wang GB, Li ZQ, et al. SWATH based quantitative proteomics analysis reveals Hfq2 play an important role on pleiotropic physiological functions in Aeromonas hydrophila[J]. J Proteomics, 2019, 195: 1-10.
doi: 10.1016/j.jprot.2018.12.030 URL |
| [16] |
Erwin S, Foster DM, Jacob ME, et al. The effect of enrofloxacin on enteric Escherichia coli: fitting a mathematical model to in vivo data[J]. PLoS One, 2020, 15(1): e0228138.
doi: 10.1371/journal.pone.0228138 URL |
| [17] |
Bush NG, Diez-Santos I, Abbott LR, et al. Quinolones: mechanism, lethality and their contributions to antibiotic resistance[J]. Molecules, 2020, 25(23): 5662.
doi: 10.3390/molecules25235662 URL |
| [18] |
Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics[J]. MedChemComm, 2019, 10(10): 1719-1739.
doi: 10.1039/c9md00120d pmid: 31803393 |
| [19] |
Li J, Wang T, Shao B, et al. Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: potential transfer to agricultural lands[J]. Environ Health Perspect, 2012, 120(8): 1144-1149.
doi: 10.1289/ehp.1104776 URL |
| [20] |
Girijan SK, Paul R, V J RK, et al. Investigating the impact of hospital antibiotic usage on aquatic environment and aquaculture systems: a molecular study of quinolone resistance in Escherichia coli[J]. Sci Total Environ, 2020, 748: 141538.
doi: 10.1016/j.scitotenv.2020.141538 URL |
| [21] |
Peng B, Su YB, Li H, et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria[J]. Cell Metab, 2015, 21(2): 249-262.
doi: S1550-4131(15)00009-1 pmid: 25651179 |
| [1] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [2] | 晏雄鹰, 王振, 王霞, 杨世辉. 微生物硫代谢与抗逆性[J]. 生物技术通报, 2023, 39(11): 150-167. |
| [3] | 赵海晴, 李耘, 梁严内, 刘哲, 任亚林, 李金娟. 联合用药对嗜水气单胞菌耐药性影响研究进展[J]. 生物技术通报, 2022, 38(6): 53-65. |
| [4] | 高雪彦, 陈林旭, 陈显轲, 庞昕, 潘登, 林建群. 嗜酸硫杆菌在工农业中的应用[J]. 生物技术通报, 2022, 38(5): 36-46. |
| [5] | 孙淑芳, 骆永丽, 李春辉, 金敏, 胥倩. UPLC-MS/MS测定小麦茎秆木质素单体交联结构的方法[J]. 生物技术通报, 2022, 38(10): 66-72. |
| [6] | 马小翔, 刘亚月, 聂影影, 黎燕媚, 王远, 薛欣怡, 洪鹏志, 张翼. 基于质谱的分子网络分析化学调控对土曲霉C23-3次生代谢产物及生物活性的影响[J]. 生物技术通报, 2021, 37(8): 95-110. |
| [7] | 陈杰豪, 缪玉佳, 梁超, 陶雨, 欧阳萍, 汪开毓, 耿毅, 石存斌, 李宁求. 山姜素对鱼源耐药性嗜水气单胞菌体外抗菌作用的研究[J]. 生物技术通报, 2021, 37(2): 103-110. |
| [8] | 张丽珊, 孙莉娜, 林镇平, 林向民. 嗜水气单胞菌非核糖体肽合成酶基因功能研究[J]. 生物技术通报, 2020, 36(4): 93-99. |
| [9] | 孟丽娜, 彭春莹, 李铁栋, 李博生. 基于蛋白质组学对螺旋藻砷胁迫响应机制的研究[J]. 生物技术通报, 2020, 36(4): 107-116. |
| [10] | 李小艳, 李泽琦, 汪玉倩, 于晶, 林镇平, 林向民. 嗜水气单胞菌acrA缺失菌株的构建及其生理功能的测定[J]. 生物技术通报, 2020, 36(11): 63-69. |
| [11] | 高云山, 刘丹丹, 徐俊林, 桑雨浓, 梁夏夏, 刘建欣, 王文彬. 嗜水气单胞菌孔蛋白OmpF重组表达及其免疫原性 分析[J]. 生物技术通报, 2019, 35(9): 234-243. |
| [12] | 毛然然, 李小艳, 武瑶, 张丽珊, 林镇平, 林向民. 嗜水气单胞菌OprM蛋白的克隆表达与免疫保护作用评价[J]. 生物技术通报, 2019, 35(9): 244-248. |
| [13] | 黄放,林向民. 嗜水气单胞菌bamA、bamB、bamD突变株的构建及其对外膜蛋白转运的影响[J]. 生物技术通报, 2018, 34(5): 148-153. |
| [14] | 林金杏, 杨迟, 冯丽萍, 胡建华. 斑马鱼嗜水气单胞菌的鉴定和人工感染组织病理学研究[J]. 生物技术通报, 2016, 32(9): 239-245. |
| [15] | 鲍海燕, 谷朋娟, 张翼, 穆军, 冯妍, 赵辰燕. 海洋真菌提取物损伤大肠杆菌DNA活性的筛选及活性菌株研究[J]. 生物技术通报, 2015, 31(8): 132-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||