








生物技术通报 ›› 2024, Vol. 40 ›› Issue (1): 332-343.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0492
张进威1,2,3( ), 吴元霞1,4, 孙静1,2,3, 李晓开1,3,5, 陆路5, 李周权4, 葛良鹏1,2,3(
), 吴元霞1,4, 孙静1,2,3, 李晓开1,3,5, 陆路5, 李周权4, 葛良鹏1,2,3( )
)
收稿日期:2023-05-21
出版日期:2024-01-26
发布日期:2024-02-06
通讯作者:
葛良鹏,男,博士,研究员,研究方向:实验猪资源创新开发与利用;E-mail: geliangpeng1982@163.com作者简介:张进威,男,博士,助理研究员,研究方向:SPF猪培育与利用;E-mail: jinweizhang50@163.com
基金资助:
ZHANG Jin-wei1,2,3( ), WU Yuan-xia1,4, SUN Jing1,2,3, LI Xiao-kai1,3,5, LU Lu5, LI Zhou-quan4, GE Liang-peng1,2,3(
), WU Yuan-xia1,4, SUN Jing1,2,3, LI Xiao-kai1,3,5, LU Lu5, LI Zhou-quan4, GE Liang-peng1,2,3( )
)
Received:2023-05-21
Published:2024-01-26
Online:2024-02-06
摘要:
【目的】以猪为动物模型研究共生微生物对宿主肠道发育代谢的调控作用。【方法】经无菌剖腹产、无菌饲养等技术培育无菌(germ-free, GF)仔猪和无特定病原(specific pathogen-free, SPF)仔猪,通过形态学观察、液相色谱分析、RNA-seq等方法研究共生微生物对仔猪肠道形态、代谢、基因表达及线粒体功能的影响。【结果】共生微生物对仔猪肠道的形态结构、短链脂肪酸含量、氨基酸代谢和肠道细胞线粒体含量等方面产生不同程度的影响;共生微生物影响仔猪肝脏、回肠和结肠组织的整体基因表达,调控线粒体氧化磷酸化、氨基酸代谢、脂质代谢等生物学过程相关基因表达,导致线粒体功能发生改变,从而影响仔猪肠道营养物质的吸收与代谢。【结论】共生微生物通过调控肠道细胞线粒体从而影响仔猪肠道发育和代谢,为“微生物-宿主互作”调控仔猪肠道健康相关研究奠定理论基础。
张进威, 吴元霞, 孙静, 李晓开, 陆路, 李周权, 葛良鹏. 共生微生物对仔猪肠道发育、代谢和线粒体功能的影响[J]. 生物技术通报, 2024, 40(1): 332-343.
ZHANG Jin-wei, WU Yuan-xia, SUN Jing, LI Xiao-kai, LU Lu, LI Zhou-quan, GE Liang-peng. Effects of Commensal Microbiota on Intestinal Development, Metabolism, and Mitochondrial Function in Piglets[J]. Biotechnology Bulletin, 2024, 40(1): 332-343.
| 目的基因 Target gene | 引物序列Primer sequence(5'-3') | 登录号Accession No. | 产物长度Product length/bp |
|---|---|---|---|
| p53 | Forward: ATTTCACCCTCCAGATCCGTG | NM_213824.3 | 153 |
| Reverse: AGGGAGACTGCCCCTTCTTA | |||
| PIG3 | Forward: GCGGACTTACTCCAGAGACAA | XM_003125363.6 | 194 |
| Reverse: AGGAACCCTTCAGGGACAGT | |||
| Cyt C | Forward: GCCAACAAGAACAAAGGCATC | NM_001129970.1 | 113 |
| Reverse: CTCCCTTCTTCTTAATGCCAGC | |||
| Apaf-1 | Forward: TACCCTGTTGGCGACTGGAGATG | XM_021093026.1 | 87 |
| Reverse: ACTGGAGCACACGAATGAAGAAGC | |||
| NRF1 | Forward: GCCAGTGAGATGAAGAGAAACG | AK393002.1 | 166 |
| Reverse: CTACAGCAGGGACCAAAGTTCAC | |||
| TFAM | Forward: GCTCTCCGTTCAGTTTTGCG | NM_001130211.1 | 187 |
| Reverse: GGAAGTTCCCTCCACAGCTC | |||
| PGC-1α | Forward: TGGACTGACATCGAGTGTGCT | NM_213963.2 | 127 |
| Reverse: TGAGTCCACCCAGAAAGCTG | |||
| HO-1 | Forward: TCCTGCTCAACATTCAGCTGTT | NM_001004027.1 | 135 |
| Reverse: TTGTCACGGGAGTGGAGTCT | |||
| CPT-1a | Forward: CAAGATGGGCATGAACGCTG | NM_001129805.1 | 145 |
| Reverse: TGGAATGTTGGGGTTGGTGT | |||
| ACC1 | Forward: CTGGAGGTGTATGTGCGAAG | XM_021066238.1 | 177 |
| Reverse: GTGGTTGAGGTTGGAGGAGA | |||
| GAPDH | Forward: ACATGGCCTCCAAGGAGTAAGA | NM_001206359.1 | 106 |
| Reverse: GATCGAGTTGGGGCTGTGACT |
表1 RT-qPCR引物序列
Table 1 Primer sequences used for RT-qPCR
| 目的基因 Target gene | 引物序列Primer sequence(5'-3') | 登录号Accession No. | 产物长度Product length/bp |
|---|---|---|---|
| p53 | Forward: ATTTCACCCTCCAGATCCGTG | NM_213824.3 | 153 |
| Reverse: AGGGAGACTGCCCCTTCTTA | |||
| PIG3 | Forward: GCGGACTTACTCCAGAGACAA | XM_003125363.6 | 194 |
| Reverse: AGGAACCCTTCAGGGACAGT | |||
| Cyt C | Forward: GCCAACAAGAACAAAGGCATC | NM_001129970.1 | 113 |
| Reverse: CTCCCTTCTTCTTAATGCCAGC | |||
| Apaf-1 | Forward: TACCCTGTTGGCGACTGGAGATG | XM_021093026.1 | 87 |
| Reverse: ACTGGAGCACACGAATGAAGAAGC | |||
| NRF1 | Forward: GCCAGTGAGATGAAGAGAAACG | AK393002.1 | 166 |
| Reverse: CTACAGCAGGGACCAAAGTTCAC | |||
| TFAM | Forward: GCTCTCCGTTCAGTTTTGCG | NM_001130211.1 | 187 |
| Reverse: GGAAGTTCCCTCCACAGCTC | |||
| PGC-1α | Forward: TGGACTGACATCGAGTGTGCT | NM_213963.2 | 127 |
| Reverse: TGAGTCCACCCAGAAAGCTG | |||
| HO-1 | Forward: TCCTGCTCAACATTCAGCTGTT | NM_001004027.1 | 135 |
| Reverse: TTGTCACGGGAGTGGAGTCT | |||
| CPT-1a | Forward: CAAGATGGGCATGAACGCTG | NM_001129805.1 | 145 |
| Reverse: TGGAATGTTGGGGTTGGTGT | |||
| ACC1 | Forward: CTGGAGGTGTATGTGCGAAG | XM_021066238.1 | 177 |
| Reverse: GTGGTTGAGGTTGGAGGAGA | |||
| GAPDH | Forward: ACATGGCCTCCAAGGAGTAAGA | NM_001206359.1 | 106 |
| Reverse: GATCGAGTTGGGGCTGTGACT |

图1 共生微生物对肠道绒毛高度、隐窝深度及黏膜层厚度的影响 A:回肠绒毛高度;B:回肠隐窝深度;C:回肠黏膜层厚度;D:结肠绒毛高度;E:结肠隐窝深度;F:结肠黏膜层厚度。*表示P < 0.05,**表示P < 0.01,***表示P < 0.001。下同
Fig. 1 Effects of commensal microbiota on intestinal villus height, crypt depth, and mucosal thickness A: Ileal villus height; B: ileal crypt depth; C: ileal mucosal thickness; D: colonic villus height; E: colonic crypt depth; F: colonic mucosal thickness. * indicates P < 0.05, ** P < 0.01, and *** P < 0.001. The same below
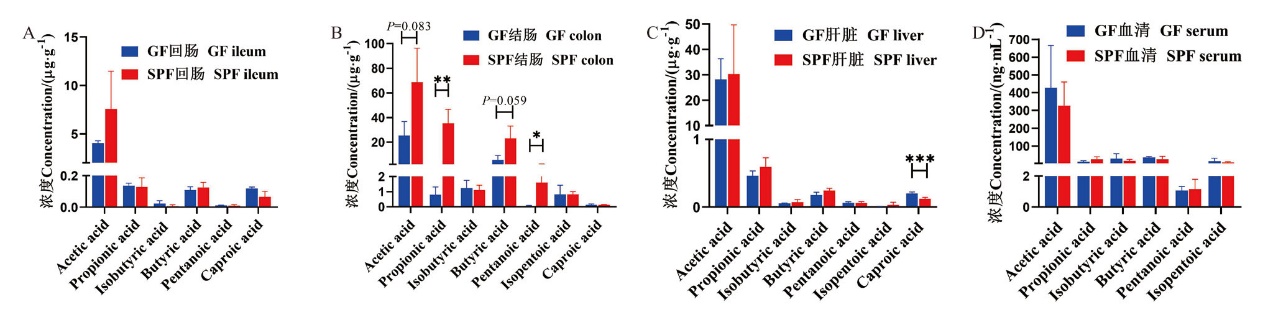
图2 共生微生物对仔猪短链脂肪酸含量的影响 A:回肠短链脂肪酸含量;B:结肠短链脂肪酸含量;C:肝脏短链脂肪酸含量;D:血清短链脂肪酸含量。Acetic acid:乙酸;Propionic acid:丙酸;Isobutyric acid:异丁酸;Butyric acid:丁酸;Pentanoic acid:戊酸;Isopentoic acid:异戊酸;Caproic acid:己酸
Fig. 2 Effects of commensal microbiota on the contents of short-chain fatty acids in piglets A: Ileum short-chain fatty acid content; B: colonic short-chain fatty acid content; C: liver short-chain fatty acid content; D: serum short-chain fatty acid content

图3 共生微生物对仔猪氨基酸含量的影响 A-B:回肠氨基酸含量;C-D:结肠氨基酸含量;E-F:肝脏氨基酸含量;G-F:血清氨基酸含量。Ala:丙氨酸;Arg:精氨酸;Asn:天冬酰胺;Asp:天冬氨酸;Cys:半胱氨酸;Gln:谷氨酰胺;Glu:谷氨酸;Gly:甘氨酸;His:组氨酸;Ile:异亮氨酸;Leu:亮氨酸;Lys:赖氨酸;Met:甲硫氨酸(蛋氨酸);Phe:苯丙氨酸;Pro:脯氨酸;Ser:丝氨酸;Thr:苏氨酸;Trp:色氨酸;Tyr:酪氨酸;Val:缬氨酸
Fig. 3 Effects of commensal microbiota on the amino acid contents in piglets A-B: Ileum amino acids content; C-D: colonic amino acids content; E-F: liver amino acids content; G-F: serum amino acids content. Ala: alanine; Arg: arginine; Asn: asparagine; Asp: asparticacid; Cys: cysteine; Gln: glutamine; Glu: glutamicacid; Gly: glycine; His: histidine; Ile: isoleucine; Leu: leucine; Lys: lysine; Met: methionine; Phe: phenylalanine; Pro: proline; Ser: serine; Thr: threonine; Trp: tryptophan; Tyr: tyrosine; Val: valine

图4 共生微生物对仔猪回肠和结肠线粒体含量的影响 左边图为线粒体免疫组化染色图片。A:回肠线粒体含量;B:结肠线粒体含量
Fig. 4 Effects of commensal microbiota on the mitochondrial contents in the ileum and colon of piglets Left picture shows mitochondria immunohistochemical staining pictures. A: Ileal mitochondrial content; B: colonic mitochondrial content
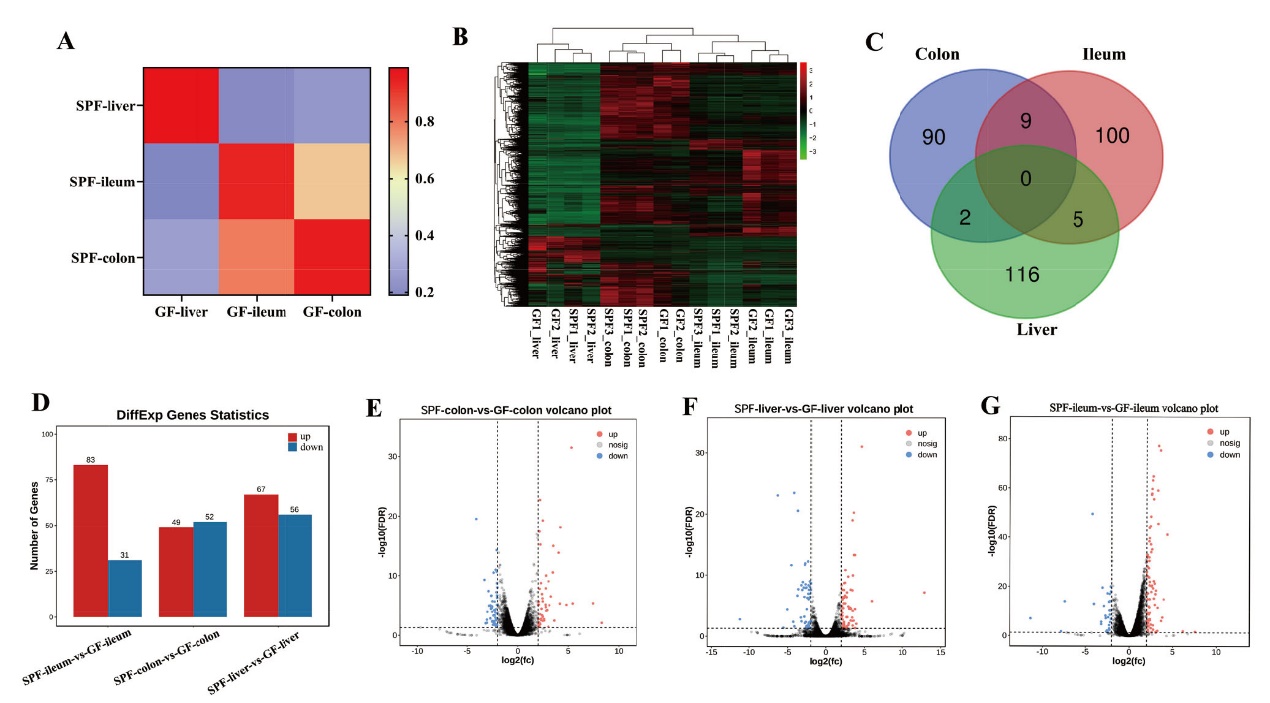
图5 共生微生物对回肠、结肠和肝脏基因表达量的影响 A:基因表达相关性分析;B:基于所有样本中mRNA整体表达的聚类分析;C:肝脏、回肠和结肠中共有和特有的差异mRNA;D-G:共生微生物对肝脏、回肠和结肠基因表达的影响
Fig. 5 Effects of commensal microbiota on the gene expressions in ileum, colon, and liver A: Gene expression correlation analysis. B: Cluster analysis based on overall gene expression in all samples. C: The common and unique differential mRNA in the liver, ileum, and colon. D-G: Commensal microbiota induced the differential gene expression in liver, ileum, and colon, respectively
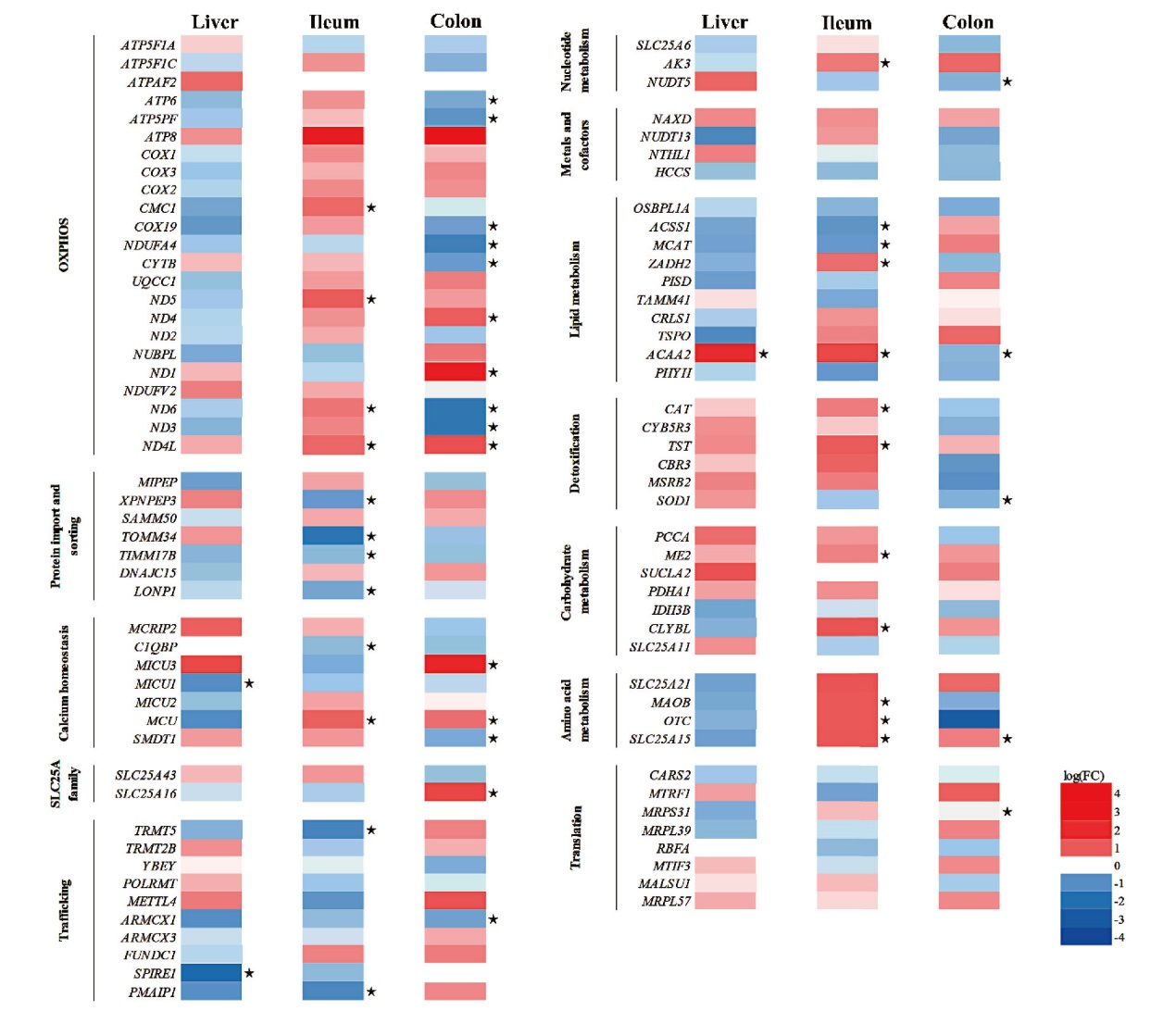
图7 共生微生物对线粒体功能相关基因表达的影响 红色为上调,蓝色为下调,★表示P < 0.05
Fig. 7 Effect of commensal microbiota on mitochondrial function-related gene expression Red indicates upregulation, blue indicates downregulation, and ★ indicates P<0.05

图8 共生微生物对线粒体功能相关基因表达的影响 A-C:肝脏脂质代谢、细胞凋亡和氧化应激过程相关基因表达量;D-F:回肠脂质代谢、细胞凋亡和氧化应激过程相关基因表达量;G-I:结肠脂质代谢、细胞凋亡和氧化应激过程相关基因表达量
Fig. 8 Effects of commensal microbiota on the expressions of mitochondrial function-related genes A-C: The expressions of genes related to lipid metabolism, apoptosis, and oxidative stress in liver. D-F: The expression of genes related to lipid metabolism, apoptosis, and oxidative stress in ileal. G-I: The expression of genes related to lipid metabolism, apoptosis and oxidative stress in colon
| [1] |
Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals[J]. Nat Immunol, 2013, 14(7): 646-653.
doi: 10.1038/ni.2604 pmid: 23778791 |
| [2] |
Rath E, Moschetta A, Haller D. Mitochondrial function - gatekeeper of intestinal epithelial cell homeostasis[J]. Nat Rev Gastroenterol Hepatol, 2018, 15(8): 497-516.
doi: 10.1038/s41575-018-0021-x pmid: 29844587 |
| [3] | Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria[J]. Genome Biol, 2001, 2(6): REVIEWS1018. |
| [4] | Franco-Obregón A, Gilbert JA. The microbiome-mitochondrion connection: common ancestries, common mechanisms, common goals[J]. mSystems, 2017, 2(3): e00018-e00017. |
| [5] |
Wang ZY, Yang ZH, Liu J, et al. Potential health benefits of whole grains: modulation of mitochondrial biogenesis and energy metabolism[J]. J Agric Food Chem, 2021, 69(47): 14065-14074.
doi: 10.1021/acs.jafc.1c05527 URL |
| [6] |
Vezza T, Abad-Jiménez Z, Marti-Cabrera M, et al. Microbiota-mitochondria inter-talk: a potential therapeutic strategy in obesity and type 2 diabetes[J]. Antioxidants, 2020, 9(9): 848.
doi: 10.3390/antiox9090848 URL |
| [7] |
Saint-Georges-Chaumet Y, Edeas M. Microbiota-mitochondria inter-talk: consequence for microbiota-host interaction[J]. Pathog Dis, 2016, 74(1): ftv096.
doi: 10.1093/femspd/ftv096 URL |
| [8] |
Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota[J]. Science, 2018, 362(6418): eaat9076.
doi: 10.1126/science.aat9076 URL |
| [9] |
Yardeni T, Tanes CE, Bittinger K, et al. Host mitochondria influence gut microbiome diversity: a role for ROS[J]. Sci Signal, 2019, 12(588): eaaw3159.
doi: 10.1126/scisignal.aaw3159 URL |
| [10] |
Duan CY, Kuang L, Xiang XM, et al. Activated Drp1-mediated mitochondrial ROS influence the gut microbiome and intestinal barrier after hemorrhagic shock[J]. Aging, 2020, 12(2): 1397-1416.
doi: 10.18632/aging.v12i2 URL |
| [11] |
Clark A, Mach N. The crosstalk between the gut microbiota and mitochondria during exercise[J]. Front Physiol, 2017, 8: 319.
doi: 10.3389/fphys.2017.00319 pmid: 28579962 |
| [12] |
Sun J, Zhong H, Du L, et al. Gene expression profiles of germ-free and conventional piglets from the same litter[J]. Sci Rep, 2018, 8(1): 10745.
doi: 10.1038/s41598-018-29093-3 pmid: 30013139 |
| [13] | 孙静, 杜蕾, 丁玉春, 等. 无菌猪的制备与微生物质量控制[J]. 中国实验动物学报, 2017, 25(6): 699-702. |
| Sun J, Du L, Ding YC, et al. Breeding and microbiological quality control of germ-free pigs[J]. Acta Lab Animalis Sci Sin, 2017, 25(6): 699-702. | |
| [14] |
Zhang JW, Shen Y, Yang GT, et al. Commensal microbiota modulates phenotypic characteristics and gene expression in piglet Peyer's patches[J]. Front Physiol, 2023, 14: 1084332.
doi: 10.3389/fphys.2023.1084332 URL |
| [15] | 胡杰. 几种物质对早期断奶仔猪生长性能、肠道结构与功能及血液指标的影响[D]. 武汉: 华中农业大学, 2004. |
| Hu J. Effects of several substances on growth performance, intestinal structure and function and blood measurements in early weaned piglets[D]. Wuhan: Huazhong Agricultural University, 2004. | |
| [16] |
Varghese F, Bukhari AB, Malhotra R, et al. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples[J]. PLoS One, 2014, 9(5): e96801.
doi: 10.1371/journal.pone.0096801 URL |
| [17] |
Seyed Jafari SM, Hunger RE. IHC optical density score: a new practical method for quantitative immunohistochemistry image analysis[J]. Appl Immunohistochem Mol Morphol, 2017, 25(1): e12-e13.
doi: 10.1097/PAI.0000000000000370 pmid: 27093452 |
| [18] |
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data[J]. Bioinformatics, 2010, 26(1): 139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [19] |
Zhou YY, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets[J]. Nat Commun, 2019, 10(1): 1523.
doi: 10.1038/s41467-019-09234-6 pmid: 30944313 |
| [20] |
Rath S, Sharma R, Gupta R, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations[J]. Nucleic Acids Res, 2021, 49(D1): D1541-D1547.
doi: 10.1093/nar/gkaa1011 pmid: 33174596 |
| [21] | 王恬, 钟翔. 仔猪肠黏膜营养与肠道修复[J]. 饲料工业, 2008, 29(2): 2-7. |
| Wang T, Zhong X. Intestines mucous membrane nutrition and intestinal damage and reparation in piglet[J]. Feed Ind, 2008, 29(2): 2-7. | |
| [22] |
Yu DF, Zhu WY, Hang SQ. Effects of long-term dietary protein restriction on intestinal morphology, digestive enzymes, gut hormones, and colonic microbiota in pigs[J]. Animals, 2019, 9(4): 180.
doi: 10.3390/ani9040180 URL |
| [23] | Roura E, Müller M, Campbell R G, et al. Digestive physiology and nutrition of swine[M]. Sustainable Swine Nutrition. 2022: 1-36. |
| [24] |
Du W, Xu H, Mei X, et al. Probiotic Bacillus enhance the intestinal epithelial cell barrier and immune function of piglets[J]. Benef Microbes, 2018, 9(5): 743-754.
doi: 10.3920/BM2017.0142 pmid: 30099892 |
| [25] |
Cummings JH, Pomare EW, Branch WJ, et al. Short chain fatty acids in human large intestine, portal, hepatic and venous blood[J]. Gut, 1987, 28(10): 1221-1227.
doi: 10.1136/gut.28.10.1221 pmid: 3678950 |
| [26] |
Morowitz MJ, Carlisle EM, Alverdy JC. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill[J]. Surg Clin North Am, 2011, 91(4): 771-785, viii.
doi: 10.1016/j.suc.2011.05.001 pmid: 21787967 |
| [27] |
Metges CC. Contribution of microbial amino acids to amino acid homeostasis of the host[J]. J Nutr, 2000, 130(7): 1857S-1864S.
doi: 10.1093/jn/130.7.1857S pmid: 10867063 |
| [28] |
Bergen WG, Wu GY. Intestinal nitrogen recycling and utilization in health and disease[J]. J Nutr, 2009, 139(5): 821-825.
doi: 10.3945/jn.109.104497 pmid: 19282369 |
| [29] |
Neis EPJG, Dejong CHC, Rensen SS. The role of microbial amino acid metabolism in host metabolism[J]. Nutrients, 2015, 7(4): 2930-2946.
doi: 10.3390/nu7042930 pmid: 25894657 |
| [30] |
Bertolini MS, Docampo R. MICU1 and MICU2 potentiation of Ca2+ uptake by the mitochondrial Ca2+ uniporter of Trypanosoma cruzi and its inhibition by Mg2[J]. Cell Calcium, 2022, 107: 102654.
doi: 10.1016/j.ceca.2022.102654 URL |
| [31] |
Giorgi C, Romagnoli A, Pinton P, et al. Ca2+ signaling, mitochondria and cell death[J]. Curr Mol Med, 2008, 8(2): 119-130.
doi: 10.2174/156652408783769571 URL |
| [32] |
Fu ZD, Selwyn FP, Cui JY, et al. RNA-Seq unveiled section-specific host response to lack of gut microbiota in mouse intestine[J]. Toxicol Appl Pharmacol, 2021, 433: 115775.
doi: 10.1016/j.taap.2021.115775 URL |
| [33] |
Zhang YN, Wang YJ, Wang XY, et al. Acetyl-coenzyme A acyltransferase 2 promote the differentiation of sheep precursor adipocytes into adipocytes[J]. J Cell Biochem, 2019, 120(5): 8021-8031.
doi: 10.1002/jcb.28080 |
| [1] | 马学虎, 马莉花, 苟妍, 马燕芬. 线粒体功能障碍引起的相关炎性疾病及靶向治疗[J]. 生物技术通报, 2023, 39(6): 119-125. |
| [2] | 王争艳, 胡海生, 雍晗紫, 鲁玉杰. 共生菌与昆虫的营养互作[J]. 生物技术通报, 2022, 38(7): 99-108. |
| [3] | 顾阳, 谭海, 员林娜, 孙海彦, 常景玲, 李志刚. 氟化钠促进节杆菌发酵合成环磷酸腺苷的生理机制[J]. 生物技术通报, 2021, 37(5): 108-116. |
| [4] | 李梦颖, 周华, 丁玉春, 刘作华, 孙静, 李周权. 肠道微生物对仔猪胆汁酸谱及胆汁酸代谢的影响[J]. 生物技术通报, 2020, 36(10): 49-61. |
| [5] | 解文雅, 邹世颖, 高如心, 贺晓云. 线粒体丙酮酸转运载体(MPC)研究进展[J]. 生物技术通报, 2019, 35(7): 196-201. |
| [6] | 张婧柔, 邵贵芳, 王姣, 张水, 杨婷玉, 邓明华. 辣椒胞质雄性不育系线粒体基因CaATP9的克隆与表达[J]. 生物技术通报, 2019, 35(11): 9-15. |
| [7] | 陈珂, 丁艳平, 王建林, 邵宝平. p53参与代谢调控的研究进展[J]. 生物技术通报, 2016, 32(11): 52-58. |
| [8] | 杨健;朱红;任碧轩;. 效应分子Map、EspF在致病性大肠杆菌致细胞线粒体功能障碍中的作用[J]. , 2009, 0(10): 169-172. |
| [9] | . 细胞工程[J]. , 1991, 0(03): 42-50. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||