








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 90-98.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0118
收稿日期:2024-01-03
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
祝诚,男,博士,副教授,研究方向:蛋白质设计;E-mail: cheng_zhu@tju.edu.cn作者简介:陈墨岩,男,硕士研究生,研究方向:蛋白质设计;E-mail: 2020226044@tju.edu.cn
Received:2024-01-03
Published:2024-07-26
Online:2024-07-30
摘要:
CRISPR/Cas12a系统能够由可编程的RNA引导,准确识别特异的单链DNA或含有PAM序列的双链DNA,而后在目标底物进行切割的同时完成对非特异性单链DNA的迅速切割,该特性使其在生物传感应用中显示出巨大前景。近年来,CRISPR/Cas12a系统被广泛应用于生物标志物的辅助检测,其生物标志物传感平台已在可视化细胞途径、体内活体诊断等生物分子传感器的构建方面得到应用。本文基于CRISPR/Cas12a生物传感平台的构建原理,对不同应用情景下CRISPR/Cas12a系统的变体进行了总结,并对基于该系统的体外、体内传感平台的应用进行了重点介绍和归纳,进一步讨论了不同传感平台的特点。最后对目前应用的优点和局限性做出总结和展望,以期为该领域的研究与应用提供一定参考。
陈墨岩, 祝诚. 基于CRISPR/Cas12a的生物传感平台的机制研究及应用[J]. 生物技术通报, 2024, 40(7): 90-98.
CHEN Mo-yan, ZHU Cheng. Mechanism Study and Application of CRISPR/Cas12a-based Biosensing Platform[J]. Biotechnology Bulletin, 2024, 40(7): 90-98.
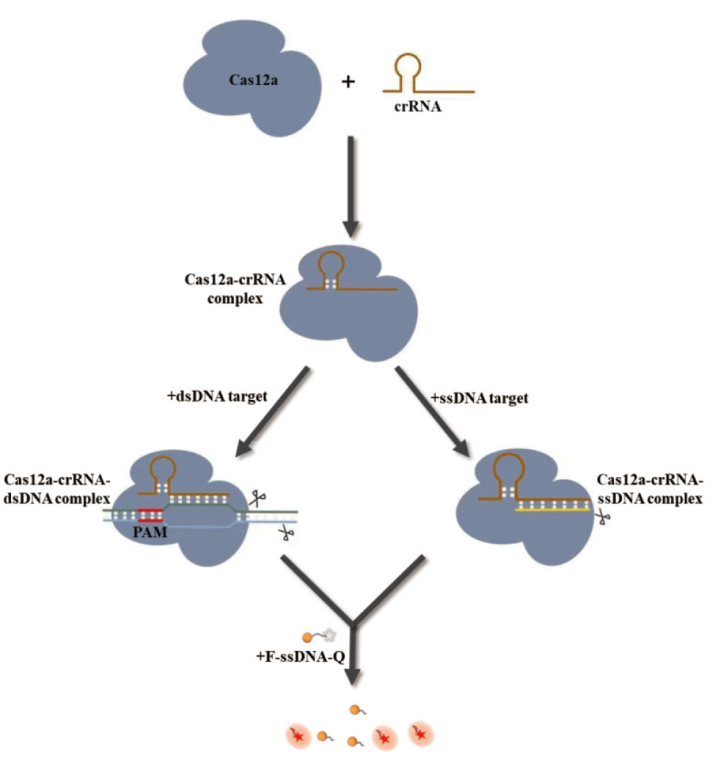
图1 CRISPR/Cas12a的原理 Cas12a-crRNA复合物在识别PAM序列后可以切割靶dsDNA(顺式)和非互补ssDNA(反式)。特异性识别crRNA的靶向ssDNA也能够激活Cas12a的顺式切割和反式切割活性,而这种活性不受PAM序列的限制
Fig. 1 Principles of CRISPR/Cas12a The Cas12a-crRNA complex cleaved target dsDNA(cis)and non-complementary ssDNA(trans)after recognizing PAM sequences. Target ssDNA that specifically recognizes crRNA is also able to activate the cis-cleavage and trans-cleavage activity of Cas12a, while the activity is not restricted by the PAM sequence

图2 基于Cas12a变体的生物传感平台 A:用于识别PAM特异性改变的变体的细菌干扰测定示意图;B:在随后的图中使用的双链断裂诱导的表达增加测定图。在框外(+3)荧光素酶基因(浅绿色)之前具有框内(+1)crRNA靶序列(橙色)的构建体稳定地整合到HEK293T细胞的基因组中。在被Cas12a/crRNA复合物切割后,非同源末端连接(NHEJ)修复将大约三分之一的下游萤光素酶基因置于框架中(深绿色),使其能够表达。11荧光素酶活性反映了Cas12a介导的切割的效率;C:后续面板中使用的目标序列列表及其前面的PAM(蓝色);D:在具有规范TTTV PAM或非规范PAM的位点上检测到的AsCas12a变体的GUIDE-seq脱靶位点的数量;E:LAMP示意图。靶基因座中的6个不同区域(F1、F2、F3、B1、B2和B3)被4个引物识别,这些引物的3' 端有一个黑色箭头,表示DNA聚合酶的延伸。FIP由一个深绿色矩形连接到一个倾斜的浅蓝色矩形来表示。BIP由一个深棕色矩形连接到一个倾斜的浅紫色矩形来表示。此外,F3置换引物用红色矩形表示,而B3置换引物用橙色矩形表示。每个区域名称后面的字母“c”表示反向互补序列。在LAMP优化过程之后,引入了群体引物,其序列相当于F1c和B1c。此外,为了证明FIP 5'端的失配如何影响LAMP反应,添加了一个黄色星号来跟踪失配的程度;F:红色箭头表示测试带,绿色箭头表示控制带
Fig. 2 A Biosensing platform based on Cas12a variant A: Schematic of bacterial interference assay used to identify variants with altered PAM specificity. B: A diagram illustrating the double-stranded-break-induced gain-of-expression assay used in subsequent figures. A construct with an in-frame(+1)crRNA target sequence(orange)preceding an out-of-frame(+3)luciferase gene(light green)is stably integrated into the genomes of HEK293T cells. Upon cleavage by a Cas12a/crRNA complex, non-homologous end-joining(NHEJ)repair places approximately one-third of the downstream luciferase genes in frame(dark green), enabling their expressions. The activities of 11 luciferase reflects the efficiency of Cas12a-mediated cleavage. C: A list of target sequences used in subsequent panels with their preceding PAMs(blue). D: Histograms illustrating the number of GUIDE-seq detected off-target sites for AsCas12a variants on sites with canonical TTTV PAMs or non-canonical PAMs. E: Schematic of LAMP. Six distinct regions(F1, F2, F3, B1, B2, and B3)in the target locus are recognized by four core primers, which have a black arrow at their 3' ends to indicate extension by the DNA polymerase. FIP is indicated by a dark green rectangle joined to a slanted light blue rectangle. BIP is indicated by a dark brown rectangle joined to a slanted light purple rectangle. In addition, the F3 displacement primer is indicated by a red rectangle, while the B3 displacement primer is indicated by an orange rectangle. The letter “c” appended to each region name indicates the reverse complementary sequence. After LAMP optimization process, writter incorporated swarm primers, whose sequences are equivalent to F1c and B1c. Moreover, to demonstrate how a mismatch at the 5' end of FIP can affect the LAMP reaction, add a yellow asterisk to track the progression of the mismatch. F: The red arrow indicates the test bands, while the green arrow indicates the control bands. Ratios of test band intensity to control band intensity are given under each dipstick
| 检测样品 Detection sample | LOD | 组合反应方法 Combined reaction method | 信号输出方式 Signal output manner | 时间 Time |
|---|---|---|---|---|
| DNA病毒/RNA病毒 DNA viruses/RNA viruses[ | 10-18 mol/L | PCR | 荧光读数 Fluorescent readout | 1 h |
| 人乳头瘤病毒 Human papillomavirus[ | 10-18 mol/L | RPA | 荧光读数 Fluorescent readout | 1 h |
| 葡萄红色斑点病毒 Grapevine red-blotch virus[ | 10-17 mol/L | PCR | 视觉比色读数 Visual colorimetric readout(aunp) | 45 min |
| 沙门氏菌Salmonella[ | 1 CFU/mL | RPA | 视觉色度读数 Visual colorimetric readout(TMB) | 3 h |
| 人乳头瘤病毒 Human papillomavirus[ | 5×10-11 mol/L | / | 电化学读数 Electrochemical readout | 1 h |
| 大肠杆菌O157:H7 Escherichia coli O157:H7[ | 19 CFU/mL | Aptamer | ||
| 非洲猪瘟病毒 African swine fever virus[ | 20 copies | RAA | 横向流动条读数 Lateral flow strip readout | 1 h |
| 大鼠肉瘤病毒癌基因Kirsten rat sarcoma viral oncogene[ | 2.6×10-11 mol/L | Poly-invertase-DNA immobilized magnetic Beads | 便携式血糖仪读数 Portable glucose meter readout | 2 h |
| 心肌钙蛋白 Cardiac troponins I[ | 7.5 ng/mL | |||
| SARS冠状病毒2型 SARS-CoV-2[ | 1.6×10-18 mol/L | RPA | 妊娠试纸条读数 Pregnancy test strip readout | 30 min |
| 尿酸 Uric acid[ | 10-8 mol/L | Allosteric transcription Factors | 荧光读数 Fluorescent readout | 25 min |
| 对羟基苯甲酸 P-hydroxybenzoic acid[ | 1.8×10-9 mol/L | |||
| 胞外体 Exosome[ | 103 particles/μL | CD63 aptamer | 荧光读数 Fluorescent readout | 2 h |
| ATP[ | 4.75×10-6 mol/L | Aptamer | 便携式荧光计读数 Portable fluorimeter readout | 15 min |
| Na+[ | 10-10 mol/L |
表1 基于CRISPR/Cas12a的生物传感策略
Table 1 CRISPR/Cas12a-based biosensing strategy
| 检测样品 Detection sample | LOD | 组合反应方法 Combined reaction method | 信号输出方式 Signal output manner | 时间 Time |
|---|---|---|---|---|
| DNA病毒/RNA病毒 DNA viruses/RNA viruses[ | 10-18 mol/L | PCR | 荧光读数 Fluorescent readout | 1 h |
| 人乳头瘤病毒 Human papillomavirus[ | 10-18 mol/L | RPA | 荧光读数 Fluorescent readout | 1 h |
| 葡萄红色斑点病毒 Grapevine red-blotch virus[ | 10-17 mol/L | PCR | 视觉比色读数 Visual colorimetric readout(aunp) | 45 min |
| 沙门氏菌Salmonella[ | 1 CFU/mL | RPA | 视觉色度读数 Visual colorimetric readout(TMB) | 3 h |
| 人乳头瘤病毒 Human papillomavirus[ | 5×10-11 mol/L | / | 电化学读数 Electrochemical readout | 1 h |
| 大肠杆菌O157:H7 Escherichia coli O157:H7[ | 19 CFU/mL | Aptamer | ||
| 非洲猪瘟病毒 African swine fever virus[ | 20 copies | RAA | 横向流动条读数 Lateral flow strip readout | 1 h |
| 大鼠肉瘤病毒癌基因Kirsten rat sarcoma viral oncogene[ | 2.6×10-11 mol/L | Poly-invertase-DNA immobilized magnetic Beads | 便携式血糖仪读数 Portable glucose meter readout | 2 h |
| 心肌钙蛋白 Cardiac troponins I[ | 7.5 ng/mL | |||
| SARS冠状病毒2型 SARS-CoV-2[ | 1.6×10-18 mol/L | RPA | 妊娠试纸条读数 Pregnancy test strip readout | 30 min |
| 尿酸 Uric acid[ | 10-8 mol/L | Allosteric transcription Factors | 荧光读数 Fluorescent readout | 25 min |
| 对羟基苯甲酸 P-hydroxybenzoic acid[ | 1.8×10-9 mol/L | |||
| 胞外体 Exosome[ | 103 particles/μL | CD63 aptamer | 荧光读数 Fluorescent readout | 2 h |
| ATP[ | 4.75×10-6 mol/L | Aptamer | 便携式荧光计读数 Portable fluorimeter readout | 15 min |
| Na+[ | 10-10 mol/L |
| [1] |
Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. J Bacteriol, 1987, 169(12): 5429-5433.
doi: 10.1128/jb.169.12.5429-5433.1987 pmid: 3316184 |
| [2] |
Nuñez JK, Kranzusch PJ, Noeske J, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity[J]. Nat Struct Mol Biol, 2014, 21(6): 528-534.
doi: 10.1038/nsmb.2820 pmid: 24793649 |
| [3] |
Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and Archaea[J]. Annu Rev Biochem, 2013, 82: 237-266.
doi: 10.1146/annurev-biochem-072911-172315 pmid: 23495939 |
| [4] |
Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and Archaea[J]. Science, 2010, 327(5962): 167-170.
doi: 10.1126/science.1179555 pmid: 20056882 |
| [5] | Garneau JE, Dupuis MÈ, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA[J]. Nature, 2010, 468(7320): 67-71. |
| [6] |
Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems[J]. Nat Rev Microbiol, 2011, 9(6): 467-477.
doi: 10.1038/nrmicro2577 pmid: 21552286 |
| [7] | Hwang WY, Fu YF, Reyon D, et al. Heritable and precise zebrafish genome editing using a CRISPR-Cas system[J]. PLoS One, 2013, 8(7): e68708. |
| [8] |
Mahas A, Stewart CN Jr, Mahfouz MM. Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation[J]. Biotechnol Adv, 2018, 36(1): 295-310.
doi: S0734-9750(17)30150-7 pmid: 29197619 |
| [9] |
Yin YF, Wang Q, Xiao L, et al. Advances in the engineering of the gene editing enzymes and the genomes: understanding and handling the off-target effects of CRISPR/Cas9[J]. J Biomed Nanotechnol, 2018, 14(3): 456-476.
doi: 10.1166/jbn.2018.2537 pmid: 29663920 |
| [10] |
Chen JS, Ma EB, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360(6387): 436-439.
doi: 10.1126/science.aar6245 pmid: 29449511 |
| [11] | Liu YF, Xu HP, Liu C, et al. CRISPR-Cas13a nanomachine based simple technology for avian influenza A(H7N9)virus on-site detection[J]. J Biomed Nanotechnol, 2019, 15(4): 790-798. |
| [12] | Khan H, Khan A, Liu YF, et al. CRISPR-Cas13a mediated nanosystem for attomolar detection of canine parvovirus type 2[J]. Chin Chem Lett, 2019, 30(12): 2201-2204. |
| [13] | Wang Y, Peng Y, Zhou HY, et al. A universal CRISPR-Cas14a responsive triple-sensitized upconversion photoelectrochemical sensor[J]. J Nanobiotechnology, 2023, 21(1): 389. |
| [14] |
Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771.
doi: 10.1016/j.cell.2015.09.038 pmid: 26422227 |
| [15] |
Swarts DC, van der Oost J, Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a[J]. Mol Cell, 2017, 66(2): 221-233.e4.
doi: S1097-2765(17)30206-X pmid: 28431230 |
| [16] | Fonfara I, Richter H, Bratovič M, et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA[J]. Nature, 2016, 532(7600): 517-521. |
| [17] | Li SY, Cheng QX, Wang JM, et al. CRISPR-Cas12a-assisted nucleic acid detection[J]. Cell Discov, 2018, 4: 20. |
| [18] |
Gao LY, Cox DBT, Yan WX, et al. Engineered Cpf1 variants with altered PAM specificities[J]. Nat Biotechnol, 2017, 35(8): 789-792.
doi: 10.1038/nbt.3900 pmid: 28581492 |
| [19] | Tran MH, Park H, Nobles CL, et al. A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020[J]. Mol Ther Nucleic Acids, 2021, 24: 40-53. |
| [20] |
Kleinstiver BP, Sousa AA, Walton RT, et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing[J]. Nat Biotechnol, 2019, 37(3): 276-282.
doi: 10.1038/s41587-018-0011-0 pmid: 30742127 |
| [21] | Ooi KH, Liu MM, Tay JWD, et al. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing[J]. Nat Commun, 2021, 12(1): 1739. |
| [22] |
Collins SP, Rostain W, Liao CY, et al. Sequence-independent RNA sensing and DNA targeting by a split domain CRISPR-Cas12a gRNA switch[J]. Nucleic Acids Res, 2021, 49(5): 2985-2999.
doi: 10.1093/nar/gkab100 pmid: 33619539 |
| [23] |
Oesinghaus L, Simmel FC. Switching the activity of Cas12a using guide RNA strand displacement circuits[J]. Nat Commun, 2019, 10(1): 2092.
doi: 10.1038/s41467-019-09953-w pmid: 31064995 |
| [24] | Wang DX, Wang YX, Wang J, et al. MnO2 nanosheets as a carrier and accelerator for improved live-cell biosensing application of CRISPR/Cas12a[J]. Chem Sci, 2022, 13(15): 4364-4371. |
| [25] | Zhang DC, Yan YR, Cheng XX, et al. Controlling the trans-cleavage of CRISPR-Cas12a with nicked PAM: universal platform for biosensing[J]. Sens Actuat B Chem, 2022, 353: 131153. |
| [26] |
Chen SY, Wang RJ, Peng S, et al. PAM-less conditional DNA substrates leverage trans-cleavage of CRISPR-Cas12a for versatile live-cell biosensing[J]. Chem Sci, 2022, 13(7): 2011-2020.
doi: 10.1039/d1sc05558e pmid: 35308851 |
| [27] |
Li YY, Mansour H, Wang T, et al. Naked-eye detection of grapevine red-blotch viral infection using a plasmonic CRISPR Cas12a assay[J]. Anal Chem, 2019, 91(18): 11510-11513.
doi: 10.1021/acs.analchem.9b03545 pmid: 31478642 |
| [28] | Dai YF, Somoza RA, Wang L, et al. Exploring the trans-cleavage activity of CRISPR-Cas12a(cpf1)for the development of a universal electrochemical biosensor[J]. Angew Chem Int Ed Engl, 2019, 58(48): 17399-17405. |
| [29] | Chen MH, Luo R, Li SH, et al. Paper-based strip for ultrasensitive detection of OSCC-associated salivary microRNA via CRISPR/Cas12a coupling with IS-primer amplification reaction[J]. Anal Chem, 2020, 92(19): 13336-13342. |
| [30] |
Xiong Y, Zhang JJ, Yang ZL, et al. Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets[J]. J Am Chem Soc, 2020, 142(1): 207-213.
doi: 10.1021/jacs.9b09211 pmid: 31800219 |
| [31] | Kim H, Lee S, Yoon J, et al. CRISPR/Cas12a collateral cleavage activity for simple and rapid detection of protein/small molecule interaction[J]. Biosens Bioelectron, 2021, 194: 113587. |
| [32] | Liang MD, Li ZL, Wang WS, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules[J]. Nat Commun, 2019, 10(1): 3672. |
| [33] | Bu SJ, Liu X, Wang Z, et al. Ultrasensitive detection of pathogenic bacteria by CRISPR/Cas12a coupling with a primer exchange reaction[J]. Sens Actuat B Chem, 2021, 347: 130630. |
| [34] | Li Y, Deng F, Hall T, et al. CRISPR/Cas12a-powered immunosensor suitable for ultra-sensitive whole Cryptosporidium oocyst detection from water samples using a plate reader[J]. Water Res, 2021, 203: 117553. |
| [35] | Yin LJ, Duan NH, Chen S, et al. Ultrasensitive pathogenic bacteria detection by a smartphone-read G-quadruplex-based CRISPR-Cas12a bioassay[J]. Sens Actuat B Chem, 2021, 347: 130586. |
| [36] | Wang XJ, Ji PP, Fan HY, et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus[J]. Commun Biol, 2020, 3(1): 62. |
| [37] |
Liu R, He Y, Lan T, et al. Installing CRISPR-Cas12a sensors in a portable glucose meter for point-of-care detection of analytes[J]. Analyst, 2021, 146(10): 3114-3120.
doi: 10.1039/d1an00008j pmid: 33999055 |
| [38] | Tang YD, Qi LJ, Liu YC, et al. CLIPON: a CRISPR-enabled strategy that turns commercial pregnancy test strips into general point-of-need test devices[J]. Angew Chem Int Ed Engl, 2022, 61(12): e202115907. |
| [39] |
Zhao XX, Zhang WQ, Qiu XP, et al. Rapid and sensitive exosome detection with CRISPR/Cas12a[J]. Anal Bioanal Chem, 2020, 412(3): 601-609.
doi: 10.1007/s00216-019-02211-4 pmid: 31897558 |
| [40] |
Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442.
doi: 10.1126/science.aam9321 pmid: 28408723 |
| [41] | Pan YC, Luan XW, Zeng F, et al. Hollow covalent organic framework-sheltering CRISPR/Cas12a as an in-vivo nanosensor for ATP imaging[J]. Biosens Bioelectron, 2022, 209: 114239. |
| [42] |
Anzalone, Andrew V, et al. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature biotechnology, 2020, 38(7):824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [43] |
Schindele P, Puchta H. Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing[J]. Plant Biotechnol J, 2020, 18(5): 1118-1120.
doi: 10.1111/pbi.13275 pmid: 31606929 |
| [44] | Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol.[J]. 2019 ;20(8):490-507. |
| [45] |
Campa CC, Weisbach NR, Santinha AJ, et al. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts[J]. Nat Methods, 2019, 16(9): 887-893.
doi: 10.1038/s41592-019-0508-6 pmid: 31406383 |
| [46] | Griffith AL, Zheng FY, McGee AV, et al. Optimization of Cas12a for multiplexed genome-scale transcriptional activation[J]. Cell Genomics, 2023, 3(9): 100387. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
