








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 99-107.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0225
何玙冰1,2( ), 付振浩1,2, 李仁瀚1,2, 刘秀霞1,2, 刘春立1,2, 杨艳坤1,2, 李业1,2(
), 付振浩1,2, 李仁瀚1,2, 刘秀霞1,2, 刘春立1,2, 杨艳坤1,2, 李业1,2( ), 白仲虎1,2(
), 白仲虎1,2( )
)
收稿日期:2024-03-08
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
李业,男,博士,助理研究员,研究方向:微生物代谢工程、合成生物学;E-mail: yeli0622@jiangnan.edu.cn;作者简介:何玙冰,女,硕士研究生,研究方向:微生物代谢工程;E-mail: 6210201011@stu.jiangnan.edu.cn
基金资助:
HE Yu-bing1,2( ), FU Zhen-hao1,2, LI Ren-han1,2, LIU Xiu-xia1,2, LIU Chun-li1,2, YANG Yan-kun1,2, LI Ye1,2(
), FU Zhen-hao1,2, LI Ren-han1,2, LIU Xiu-xia1,2, LIU Chun-li1,2, YANG Yan-kun1,2, LI Ye1,2( ), BAI Zhong-hu1,2(
), BAI Zhong-hu1,2( )
)
Received:2024-03-08
Published:2024-07-26
Online:2024-07-30
摘要:
【目的】 为实现生物合成重要化合物2-萘乙醇,探索利于2-萘乙醇合成的基因组合。【方法】 向酿酒酵母BY4741发酵液中外源添加2-萘丙氨酸,运用高效液相色谱法进行产物检测;绘制BY4741的生长曲线和产量曲线,添加不同浓度2-萘丙氨酸到BY4741发酵液中并检测BY4741生长和生产情况;对BY4741进行代谢工程改造,构建过表达Ehrlich途径6个基因的质粒并转化进BY4741中;发酵6株改造菌株,检测产物产量。【结果】 发酵液中检测到了推测产物2-萘乙醇,代谢工程改造菌株相较于野生型BY4741菌株产量均有所提升,其中过表达ARO9和ARO10基因使得2-萘乙醇的产量在外源添加1 mmol/L 2-萘丙氨酸的情况下达到0.258 mmol/L,转化率达到野生型的2.3倍。【结论】 在酿酒酵母中实现了2-萘乙醇的生物合成,探索出ARO9和ARO10是利于2-萘乙醇生物合成的基因组合,为工业生产2-萘乙醇提供了一种新的生物合成方法。
何玙冰, 付振浩, 李仁瀚, 刘秀霞, 刘春立, 杨艳坤, 李业, 白仲虎. 利用代谢工程在酿酒酵母中高效合成2-萘乙醇[J]. 生物技术通报, 2024, 40(7): 99-107.
HE Yu-bing, FU Zhen-hao, LI Ren-han, LIU Xiu-xia, LIU Chun-li, YANG Yan-kun, LI Ye, BAI Zhong-hu. Efficient Biosynthesis of 2-Naphthaleneethanol in Metabolically Engineered Saccharomyces cerevisiae[J]. Biotechnology Bulletin, 2024, 40(7): 99-107.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| ARO8_F | GCATCGTCTCATCGGTCTCATATGACTTTACCTGAATCAAAAGACTTTTCTT |
| ARO8_R | ATGCCGTCTCAGGTCTCAGGATCCTTTGGAAATACCAAATTCTTCGTATAAAGT |
| ARO9_F | GCATCGTCTCATCGGTCTCATATGACTGCTGGTTCTGCC |
| ARO9_R | ATGCCGTCTCAGGTCTCAGGATCCACTTTTATAGTTGTCAAAAAATTCTTTTATGCC |
| ARO10_F | GCATCGTCTCATCGGTCTCATATGGCACCTGTTACAATTGAAAAGT |
| ARO10_R | ATGCCGTCTCAGGTCTCAGGATCCTTTTTTATTTCTTTTAAGTGCCGCTGC |
| PDC1_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATTACTTTGGGTAAATATTTGT |
| PDC1_R | ATGCCGTCTCAGGTCTCAGGATCCTTGCTTAGCGTTGGTAGCAG |
| PDC5_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATAACCTTAGGTAAATATTTATTTGAA |
| PDC5_R | ATGCCGTCTCAGGTCTCAGGATCCTTGTTTAGCGTTAGTAGCGGC |
| PDC6_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATTACTCTTGGAAAATACTT |
| PDC6_R | ATGCCGTCTCAGGTCTCAGGATCCTTGTTTGGCATTTGTAGCGG |
| HO5'_F | CACATCATTTTCGTGGATCC |
| HO5'_R | ACAGCGATGGAACTTACGGC |
| HO3'_F | TATCGTGTTGCATCTGCGGC |
| HO3'_R | CTTTGGACTTAAAATGGCGT |
表1 本研究所使用的引物
Table 1 Primers used in this study
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| ARO8_F | GCATCGTCTCATCGGTCTCATATGACTTTACCTGAATCAAAAGACTTTTCTT |
| ARO8_R | ATGCCGTCTCAGGTCTCAGGATCCTTTGGAAATACCAAATTCTTCGTATAAAGT |
| ARO9_F | GCATCGTCTCATCGGTCTCATATGACTGCTGGTTCTGCC |
| ARO9_R | ATGCCGTCTCAGGTCTCAGGATCCACTTTTATAGTTGTCAAAAAATTCTTTTATGCC |
| ARO10_F | GCATCGTCTCATCGGTCTCATATGGCACCTGTTACAATTGAAAAGT |
| ARO10_R | ATGCCGTCTCAGGTCTCAGGATCCTTTTTTATTTCTTTTAAGTGCCGCTGC |
| PDC1_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATTACTTTGGGTAAATATTTGT |
| PDC1_R | ATGCCGTCTCAGGTCTCAGGATCCTTGCTTAGCGTTGGTAGCAG |
| PDC5_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATAACCTTAGGTAAATATTTATTTGAA |
| PDC5_R | ATGCCGTCTCAGGTCTCAGGATCCTTGTTTAGCGTTAGTAGCGGC |
| PDC6_F | GCATCGTCTCATCGGTCTCATATGTCTGAAATTACTCTTGGAAAATACTT |
| PDC6_R | ATGCCGTCTCAGGTCTCAGGATCCTTGTTTGGCATTTGTAGCGG |
| HO5'_F | CACATCATTTTCGTGGATCC |
| HO5'_R | ACAGCGATGGAACTTACGGC |
| HO3'_F | TATCGTGTTGCATCTGCGGC |
| HO3'_R | CTTTGGACTTAAAATGGCGT |
| 名称 Name | 携带基因/特征 Characteristics of carried gene | 大肠杆菌抗性标记 E. coli marker | 部件类型/使用的部件 Type/type used | 来源 Origin |
|---|---|---|---|---|
| pYTK001 | Part plasmid entry vector | CamR | entry vector | Lee等[ |
| pYTK002 | ConLS | CamR | 1 | Lee等[ |
| pYTK003 | ConL1 | CamR | 1 | Lee等[ |
| pYTK004 | ConL2 | CamR | 1 | Lee等[ |
| pYTK009 | pTDH3 | CamR | 2 | Lee等[ |
| pYTK011 | pPGK1 | CamR | 2 | Lee等[ |
| pYTK048 | Spacer | CamR | 234 | Lee等[ |
| pYTK051 | tENO1 | CamR | 4 | Lee等[ |
| pYTK052 | tSSA1 | CamR | 4 | Lee等[ |
| pYTK067 | ConR1 | CamR | 5 | Lee等[ |
| pYTK068 | ConR2 | CamR | 5 | Lee等[ |
| pYTK072 | ConRE | CamR | 5 | Lee等[ |
| pYTK095 | AmpR-ColE1 | AmpR | 678 | Lee等[ |
| pBL602 | Integration(HO-loc)HIS3 GFP Dropout | KanR | 15678 | 实验室保藏 |
| pBL617 | ARO8 | CamR | 3 | 实验室保藏 |
| pBL618 | ARO9 | CamR | 3 | 实验室保藏 |
| pBL621 | ARO10 | CamR | 3 | 实验室保藏 |
| pBL623 | PDC1 | CamR | 3 | 实验室保藏 |
| pBL624 | PDC5 | CamR | 3 | 实验室保藏 |
| pBL625 | PDC6 | CamR | 3 | 实验室保藏 |
| pBL883 | ConL1-Spacer-ConRE | AmpR | pYTK003, pYTK048, pYTK072, pYTK095 | 本研究构建 |
| pBL884 | ConL2-Spacer-ConRE | AmpR | pYTK004, pYTK048, pYTK072, pYTK096 | 本研究构建 |
| pBL885 | ConLS-pPGK1-ARO8-tENO1-ConR1 | AmpR | pYTK002, pYTK011, pYTK051, pYTK067, pYTK095, pBL617 | 本研究构建 |
| pBL886 | ConLS-pPGK1-ARO9-tENO1-ConR1 | AmpR | pYTK002, pYTK011, pYTK051, pYTK067, pYTK095, pBL618 | 本研究构建 |
| pBL889 | ConL1-pTDH3-ARO10-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL621 | 本研究构建 |
| pBL890 | ConL1-pTDH3-PDC1-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL623 | 本研究构建 |
| pBL891 | ConL1-pTDH3-PDC5-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL624 | 本研究构建 |
| pBL892 | ConL1-pTDH3-PDC6-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL625 | 本研究构建 |
| pBL909 | Integration(HO-loc)HIS3 pPGK1-ARO8-tENO1 | KanR | pBL602, pBL883, pBL885 | 本研究构建 |
| pBL910 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 | KanR | pBL602, pBL883, pBL886 | 本研究构建 |
| pBL961 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-ARO10-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL889 | 本研究构建 |
| pBL962 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC1-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL890 | 本研究构建 |
| pBL963 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC5-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL891 | 本研究构建 |
| pBL964 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC6-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL892 | 本研究构建 |
表2 本研究所使用的质粒
Table 2 Plasmids used in this study
| 名称 Name | 携带基因/特征 Characteristics of carried gene | 大肠杆菌抗性标记 E. coli marker | 部件类型/使用的部件 Type/type used | 来源 Origin |
|---|---|---|---|---|
| pYTK001 | Part plasmid entry vector | CamR | entry vector | Lee等[ |
| pYTK002 | ConLS | CamR | 1 | Lee等[ |
| pYTK003 | ConL1 | CamR | 1 | Lee等[ |
| pYTK004 | ConL2 | CamR | 1 | Lee等[ |
| pYTK009 | pTDH3 | CamR | 2 | Lee等[ |
| pYTK011 | pPGK1 | CamR | 2 | Lee等[ |
| pYTK048 | Spacer | CamR | 234 | Lee等[ |
| pYTK051 | tENO1 | CamR | 4 | Lee等[ |
| pYTK052 | tSSA1 | CamR | 4 | Lee等[ |
| pYTK067 | ConR1 | CamR | 5 | Lee等[ |
| pYTK068 | ConR2 | CamR | 5 | Lee等[ |
| pYTK072 | ConRE | CamR | 5 | Lee等[ |
| pYTK095 | AmpR-ColE1 | AmpR | 678 | Lee等[ |
| pBL602 | Integration(HO-loc)HIS3 GFP Dropout | KanR | 15678 | 实验室保藏 |
| pBL617 | ARO8 | CamR | 3 | 实验室保藏 |
| pBL618 | ARO9 | CamR | 3 | 实验室保藏 |
| pBL621 | ARO10 | CamR | 3 | 实验室保藏 |
| pBL623 | PDC1 | CamR | 3 | 实验室保藏 |
| pBL624 | PDC5 | CamR | 3 | 实验室保藏 |
| pBL625 | PDC6 | CamR | 3 | 实验室保藏 |
| pBL883 | ConL1-Spacer-ConRE | AmpR | pYTK003, pYTK048, pYTK072, pYTK095 | 本研究构建 |
| pBL884 | ConL2-Spacer-ConRE | AmpR | pYTK004, pYTK048, pYTK072, pYTK096 | 本研究构建 |
| pBL885 | ConLS-pPGK1-ARO8-tENO1-ConR1 | AmpR | pYTK002, pYTK011, pYTK051, pYTK067, pYTK095, pBL617 | 本研究构建 |
| pBL886 | ConLS-pPGK1-ARO9-tENO1-ConR1 | AmpR | pYTK002, pYTK011, pYTK051, pYTK067, pYTK095, pBL618 | 本研究构建 |
| pBL889 | ConL1-pTDH3-ARO10-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL621 | 本研究构建 |
| pBL890 | ConL1-pTDH3-PDC1-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL623 | 本研究构建 |
| pBL891 | ConL1-pTDH3-PDC5-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL624 | 本研究构建 |
| pBL892 | ConL1-pTDH3-PDC6-tSSA1-ConR2 | AmpR | pYTK003, pYTK009, pYTK052, pYTK068, pYTK095, pBL625 | 本研究构建 |
| pBL909 | Integration(HO-loc)HIS3 pPGK1-ARO8-tENO1 | KanR | pBL602, pBL883, pBL885 | 本研究构建 |
| pBL910 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 | KanR | pBL602, pBL883, pBL886 | 本研究构建 |
| pBL961 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-ARO10-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL889 | 本研究构建 |
| pBL962 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC1-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL890 | 本研究构建 |
| pBL963 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC5-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL891 | 本研究构建 |
| pBL964 | Integration(HO-loc)HIS3 pPGK1-ARO9-tENO1 pTDH3-PDC6-tSSA1 | KanR | pBL602, pBL884, pBL886, pBL892 | 本研究构建 |
| 名称Name | 基因型Genotype | 来源Origin |
|---|---|---|
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Brachmann等[ |
| yBL_909 | [BY4741]Holoc::pBL_909 | 本研究构建 |
| yBL_910 | [BY4741]Holoc::pBL_910 | 本研究构建 |
| yBL_961 | [BY4741]Holoc::pBL_961 | 本研究构建 |
| yBL_962 | [BY4741]Holoc::pBL_962 | 本研究构建 |
| yBL_963 | [BY4741]Holoc::pBL_963 | 本研究构建 |
| yBL_964 | [BY4741]Holoc::pBL_964 | 本研究构建 |
表3 本研究使用的菌株
Table 3 Strains used in this study
| 名称Name | 基因型Genotype | 来源Origin |
|---|---|---|
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Brachmann等[ |
| yBL_909 | [BY4741]Holoc::pBL_909 | 本研究构建 |
| yBL_910 | [BY4741]Holoc::pBL_910 | 本研究构建 |
| yBL_961 | [BY4741]Holoc::pBL_961 | 本研究构建 |
| yBL_962 | [BY4741]Holoc::pBL_962 | 本研究构建 |
| yBL_963 | [BY4741]Holoc::pBL_963 | 本研究构建 |
| yBL_964 | [BY4741]Holoc::pBL_964 | 本研究构建 |
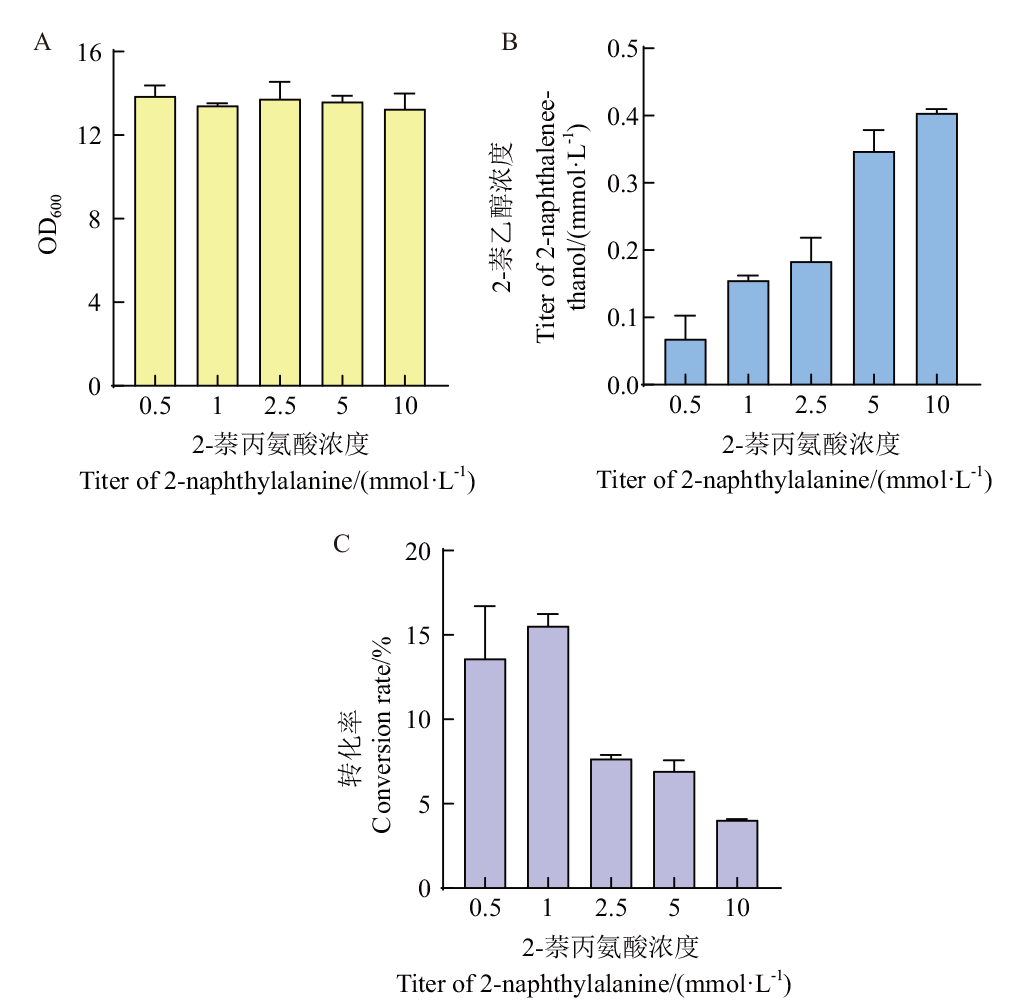
图4 BY4741生长情况(A)、2-萘乙醇生产情况(B)和转化率随着2-萘丙氨酸浓度的变化(C)
Fig. 4 Growth of BY4741(A), yield of 2-naphthaleneethanol(B)and conversion rate in different concentration of 2-naphthylalanine(C)
| [1] | Li XM, Mi QL, Gao Q, et al. Antibacterial naphthalene derivatives from the fermentation products of the endophytic fungus Phomopsis fukushii[J]. Chem Nat Compd, 2021, 57(2): 293-296. |
| [2] | 刘耀, 李枝芳, 张茂利. 萘环高分子在覆铜板中的应用[C]// 第二十届中国覆铜板技术研讨会论文集. 苏州, 2019: 228-235. |
| [3] | Jiang WN, Zhao QL, Cheng WS, et al. CuI-mediated benzannulation of(ortho-arylethynyl)phenylenaminones to assemble α-aminonaphthalene derivatives[J]. Org Chem Front, 2021, 8(13): 3250-3254. |
| [4] |
Lin B, Fan L, Ge JY, et al. A naphthalene-based fluorescent probe with a large Stokes shift for mitochondrial pH imaging[J]. Analyst, 2018, 143(20): 5054-5060.
doi: 10.1039/c8an01371c pmid: 30238115 |
| [5] | Zeng CH, Xu ZY, Song C, et al. Naphthalene-based fluorescent probe for on-site detection of hydrazine in the environment[J]. J Hazard Mater, 2023, 445: 130415. |
| [6] | Yokota K, Takeuchi S. Process for the preparation of 2-naphthylethanol, JP2006206533[P]. 2006-08-10. |
| [7] | 刘学端. 微生物生态学驱动社会与经济“绿色高效发展”[J]. 微生物学通报, 2020, 47(9): 2681-2682. |
| Liu XD. Microbial Ecology drives “Green and Efficient Development” of society and economy[J]. Microbiol China, 2020, 47(9): 2681-2682. | |
| [8] | Ehrlich F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit Dem Eiweißaufbau der Hefe[J]. Ber Dtsch Chem Ges, 1907, 40(1): 1027-1047. |
| [9] | 于爱群, 庞亚如, 胡智慧, 等. 平台化学品短链支链脂肪酸和短链支链醇的微生物代谢工程[J]. 微生物学通报, 2018, 45(1): 173-180. |
| Yu AQ, Pang YR, Hu ZH, et al. Advances in metabolic engineering for the microbial production of short branched-chain fatty acids and short branched-chain alcohols[J]. Microbiol China, 2018, 45(1): 173-180. | |
| [10] | Iraqui I, Vissers S, Cartiaux M, et al. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily[J]. Mol Gen Genet, 1998, 257(2): 238-248. |
| [11] | Vuralhan Z, Luttik MAH, Tai SL, et al. Physiological characterization of the ARO10-dependent, broad-substrate-specificity 2-oxo acid decarboxylase activity of Saccharomyces cerevisiae[J]. Appl Environ Microbiol, 2005, 71(6): 3276-3284. |
| [12] | 孙中贯, 刘琳, 王亚平, 等. 酿酒酵母高级醇代谢研究进展[J]. 生物工程学报, 2021, 37(2): 429-447. |
| Sun ZG, Liu L, Wang YP, et al. Higher alcohols metabolism by Saccharomyces cerevisiae: a mini review[J]. Chin J Biotechnol, 2021, 37(2): 429-447. | |
| [13] | Kondo T, Tezuka H, Ishii J, et al. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae[J]. J Biotechnol, 2012, 159(1/2): 32-37. |
| [14] |
孙井震, 何玙冰, 杨卫华, 等. 酿酒酵母过表达Ehrlich途径基因高效合成色醇[J]. 食品与发酵工业, 2022, 48(24): 16-23.
doi: 10.13995/j.cnki.11-1802/ts.031377 |
| Sun JZ, He YB, Yang WH, et al. Efficient synthesis of indole-3-ethanol in Saccharomyces cerevisiae by overexpression of Ehrlich pathway genes[J]. Food Ferment Ind, 2022, 48(24): 16-23. | |
| [15] | Chan WT, Verma CS, Lane DP, et al. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli[J]. Biosci Rep, 2013, 33(6): e00086. |
| [16] |
Lee ME, DeLoache WC, Cervantes B, et al. A highly characterized yeast toolkit for modular, multipart assembly[J]. ACS Synth Biol, 2015, 4(9): 975-986.
doi: 10.1021/sb500366v pmid: 25871405 |
| [17] | Casini A, Storch M, Baldwin GS, et al. Bricks and blueprints: methods and standards for DNA assembly[J]. Nat Rev Mol Cell Biol, 2015, 16(9): 568-576. |
| [18] |
Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications[J]. Yeast, 1998, 14(2): 115-132.
doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 pmid: 9483801 |
| [19] |
Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method[J]. Nat Protoc, 2007, 2(1): 31-34.
doi: 10.1038/nprot.2007.13 pmid: 17401334 |
| [20] |
Avalos JL, Fink GR, Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols[J]. Nat Biotechnol, 2013, 31(4): 335-341.
doi: 10.1038/nbt.2509 pmid: 23417095 |
| [21] | Hassing EJ, de Groot PA, Marquenie VR, et al. Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae[J]. Metab Eng, 2019, 56: 165-180. |
| [22] | Jiang JJ, Yin H, Wang S, et al. Metabolic engineering of Saccharomyces cerevisiae for high-level production of salidroside from glucose[J]. J Agric Food Chem, 2018, 66(17): 4431-4438. |
| [23] | Zhou R, Song QY, Xia HL, et al. Isolation and identification of non- Saccharomyces yeast producing 2-phenylethanol and study of the Ehrlich pathway and shikimate pathway[J]. J Fungi, 2023, 9(9): 878. |
| [24] | 刘春筱, 夏媛媛, 齐丽娜, 等. 代谢工程改造大肠杆菌合成羟基酪醇[J]. 生物工程学报, 2021, 37(12): 4243-4253. |
| Liu CX, Xia YY, Qi LN, et al. Metabolic engineering of Escherichia coli for production of hydroxytyrosol[J]. Chin J Biotechnol, 2021, 37(12): 4243-4253. | |
| [25] | Gallardo-Fernández M, Valls-Fonayet J, Valero E, et al. Isotopic labelling-based analysis elucidates biosynthesis pathways in Saccharomyces cerevisiae for Melatonin, Serotonin and Hydroxytyrosol formation[J]. Food Chem, 2022, 374: 131742. |
| [26] | Furuya T, Kino K. Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives[J]. Appl Microbiol Biotechnol, 2014, 98(3): 1145-1154. |
| [27] | Wu L, Wen YD, Chen WY, et al. Simultaneously deleting ADH2 and THI3 genes of Saccharomyces cerevisiae for reducing the yield of acetaldehyde and fusel alcohols[J]. FEMS Microbiol Lett, 2021, 368(15): fnab094. |
| [28] |
Pirkov I, Norbeck J, Gustafsson L, et al. A complete inventory of all enzymes in the eukaryotic methionine salvage pathway[J]. FEBS J, 2008, 275(16): 4111-4120.
doi: 10.1111/j.1742-4658.2008.06552.x pmid: 18625006 |
| [29] | Hazelwood LA, Daran JM, van Maris AJA, et al. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism[J]. Appl Environ Microbiol, 2008, 74(8): 2259-2266. |
| [30] | Averesch NJH, Prima A, Krömer JO. Enhanced production of para-hydroxybenzoic acid by genetically engineered Saccharomyces cerevisiae[J]. Bioprocess Biosyst Eng, 2017, 40(8): 1283-1289. |
| [31] |
Hazelwood LA, Tai SL, Boer VM, et al. A new physiological role for Pdr12p in Saccharomyces cerevisiae: export of aromatic and branched-chain organic acids produced in amino acid catabolism[J]. FEMS Yeast Res, 2006, 6(6): 937-945.
pmid: 16911515 |
| [32] |
Reifenrath M, Boles E. Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae[J]. Metab Eng, 2018, 45: 246-254.
doi: S1096-7176(17)30407-X pmid: 29330068 |
| [33] | Ishchuk OP, Domenzain I, Sánchez BJ, et al. Genome-scale modeling drives 70-fold improvement of intracellular heme production in Saccharomyces cerevisiae[J]. Proc Natl Acad Sci USA, 2022, 119(30): e2108245119. |
| [1] | 沈真辉, 曹瑶, 杨林雷, 罗祥英, 子灵山, 陆青青, 李荣春. 金耳和毛韧革菌麦角硫因生物合成基因的克隆及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 259-272. |
| [2] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [3] | 张美玉, 赵玉斌, 王灵云, 宋元达, 赵新河, 任晓洁. 微藻破囊壶菌产功能性脂肪酸DHA研究进展[J]. 生物技术通报, 2024, 40(6): 81-94. |
| [4] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [5] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [6] | 陈治民, 李翠, 韦继天, 李昕然, 刘峄, 郭强. 绿原酸生物合成调控及其应用研究进展[J]. 生物技术通报, 2024, 40(1): 57-71. |
| [7] | 何思成, 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群. 解脂耶氏酵母细胞工厂生产多不饱和脂肪酸的研究进展[J]. 生物技术通报, 2024, 40(1): 72-85. |
| [8] | 李亮, 徐姗姗, 姜艳军. 生物合成法生产麦角硫因的研究进展[J]. 生物技术通报, 2024, 40(1): 86-99. |
| [9] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [10] | 徐发迪, 徐康, 孙东明, 李萌蕾, 赵建志, 鲍晓明. 基于杨木(Populus sp.)的二代燃料乙醇技术研究进展[J]. 生物技术通报, 2023, 39(9): 27-39. |
| [11] | 程亚楠, 张文聪, 周圆, 孙雪, 李玉, 李庆刚. 乳酸乳球菌生产2'-岩藻糖基乳糖的途径构建及发酵培养基优化[J]. 生物技术通报, 2023, 39(9): 84-96. |
| [12] | 赵思佳, 王晓璐, 孙纪录, 田健, 张杰. 代谢工程改造毕赤酵母生产赤藓糖醇[J]. 生物技术通报, 2023, 39(8): 137-147. |
| [13] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [14] | 王玲, 卓燊, 付学森, 刘紫璇, 刘笑蓉, 王志辉, 周日宝, 刘湘丹. 莲生物碱生物合成途径及相关基因研究进展[J]. 生物技术通报, 2023, 39(7): 56-66. |
| [15] | 李雨真, 梅天秀, 李治文, 王淇, 李俊, 邹岳, 赵心清. 红酵母基因组和代谢工程改造研究进展[J]. 生物技术通报, 2023, 39(7): 67-79. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||