








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 129-141.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0179
杨巍1,2( ), 赵丽芬1,2,3, 唐兵1,2, 周麟笔1,2, 杨娟4, 莫传园1,2, 张宝会1,2, 李飞1,2, 阮松林5, 邓英1,2(
), 赵丽芬1,2,3, 唐兵1,2, 周麟笔1,2, 杨娟4, 莫传园1,2, 张宝会1,2, 李飞1,2, 阮松林5, 邓英1,2( )
)
收稿日期:2024-02-24
出版日期:2024-08-26
发布日期:2024-07-31
通讯作者:
邓英,女,博士,研究员,硕士生导师,研究方向:蔬菜遗传育种与栽培;E-mail: 87928883@qq.com作者简介:杨巍,男,硕士,助理研究员,研究方向:蔬菜遗传育种与栽培;E-mail: yangwei139@sina.cn
基金资助:
YANG Wei1,2( ), ZHAO Li-fen1,2,3, TANG Bing1,2, ZHOU Lin-bi1,2, YANG Juan4, MO Chuan-yuan1,2, ZHANG Bao-hui1,2, LI Fei1,2, RUAN Song-lin5, DENG Ying1,2(
), ZHAO Li-fen1,2,3, TANG Bing1,2, ZHOU Lin-bi1,2, YANG Juan4, MO Chuan-yuan1,2, ZHANG Bao-hui1,2, LI Fei1,2, RUAN Song-lin5, DENG Ying1,2( )
)
Received:2024-02-24
Published:2024-08-26
Online:2024-07-31
摘要:
【目的】SRO基因家族是植物特有的一类转录因子,在植物生长发育与胁迫响应中具有重要作用,研究芥菜SRO基因家族,为解析芥菜SRO基因功能与遗传改良提供理论依据。【方法】利用生物信息学鉴定油用芥菜与菜用芥菜基因组中的SRO基因家族成员,运用TBtools、MEGA、Cytoscape、NCBI、STRING、EggNOG等软件与数据库进行理化性质、序列特征、进化关系、调控网络等分析以及RT-qPCR分析盐胁迫下的表达模式。【结果】两种类型芥菜SRO基因家族成员数量存在差异,并分属A与B两大类,其中A类SRO蛋白含有WWE、PARP及RST结构域,而B类蛋白则缺少WWE结构域,SRO基因启动子区域含有多种非生物胁迫响应与激素响应的顺式作用元件,microRNA-BjuSRO-靶基因构建复杂调控网络,参与了细胞凋亡、根系形态建成、免疫应答、异源刺激的细胞反应、对活性氧的反应等生物学过程,油用芥菜BjuOSRO基因与甘蓝SRO基因具有较近的进化关系,RT-qPCR结果显示,在盐胁迫下BjuVA1a、BjuVA1e、BjuVA2a、BjuVA3a、BjuVB1b、BjuVA2a显著上调表达。【结论】芥菜SRO基因家族具有功能多样性,BjuVA1a、BjuVA1e、BjuVA2a、BjuVA3a、BjuVB1b、BjuVA2a与盐胁迫响应密切相关,可作为培育耐盐型芥菜新品种的候选基因。
杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141.
YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L.[J]. Biotechnology Bulletin, 2024, 40(8): 129-141.
| 基因名称Gene name | 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| BjuVA1a | BjuVA05G09830.1 | CACTAAGTCACGCCGAGGAG | CTCTCCACCCACACCAAGTC |
| BjuVA1e | BjuVA09G34200.1 | AAACATCGAGGGGACGCAAA | AATGCTCCCCCAACTCCAAG |
| BjuVA2a | BjuVB03G32070.1 | ATGTAGGCCGCTTTTCCAGT | AACGTTTGCATCCCCTCGAT |
| BjuVA3a | BjuVA08G09520.1 | ACCCAGGATCCTCAAGGTGT | CTCATCTTGCAGCTTTCCGC |
| BjuVA4a | BjuVB07G38920.1 | GCGCTTATGAGACTCGCTCT | TCGCCCTCCTCGTAGATCAT |
| BjuVB1a | BjuVB03G28110.1 | TCACAACGAGAGCCAAGCAT | AGCCAACGTACCAAGCGTAT |
| BjuVB1b | BjuVA07G14180.1 | TCGATCCACTCTCCTCCTCC | GGAGAGGAAACACGTGGTGA |
| BjuVB2a | BjuVB04G02010.1 | CGCCGTGGAGAAAACAGAGT | AGAGCATTATCCGGGGAGAGA |
| ACT7 | NM_121018.4 | GGAATCGCTGACCGTATGAG | ACCCTCCAATCCAGACACTG |
表1 BjuSROs 实时荧光定量PCR 分析所用引物
Table 1 Primer sequences used in quantitative real-time PCR analysis of BjuSROs
| 基因名称Gene name | 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| BjuVA1a | BjuVA05G09830.1 | CACTAAGTCACGCCGAGGAG | CTCTCCACCCACACCAAGTC |
| BjuVA1e | BjuVA09G34200.1 | AAACATCGAGGGGACGCAAA | AATGCTCCCCCAACTCCAAG |
| BjuVA2a | BjuVB03G32070.1 | ATGTAGGCCGCTTTTCCAGT | AACGTTTGCATCCCCTCGAT |
| BjuVA3a | BjuVA08G09520.1 | ACCCAGGATCCTCAAGGTGT | CTCATCTTGCAGCTTTCCGC |
| BjuVA4a | BjuVB07G38920.1 | GCGCTTATGAGACTCGCTCT | TCGCCCTCCTCGTAGATCAT |
| BjuVB1a | BjuVB03G28110.1 | TCACAACGAGAGCCAAGCAT | AGCCAACGTACCAAGCGTAT |
| BjuVB1b | BjuVA07G14180.1 | TCGATCCACTCTCCTCCTCC | GGAGAGGAAACACGTGGTGA |
| BjuVB2a | BjuVB04G02010.1 | CGCCGTGGAGAAAACAGAGT | AGAGCATTATCCGGGGAGAGA |
| ACT7 | NM_121018.4 | GGAATCGCTGACCGTATGAG | ACCCTCCAATCCAGACACTG |
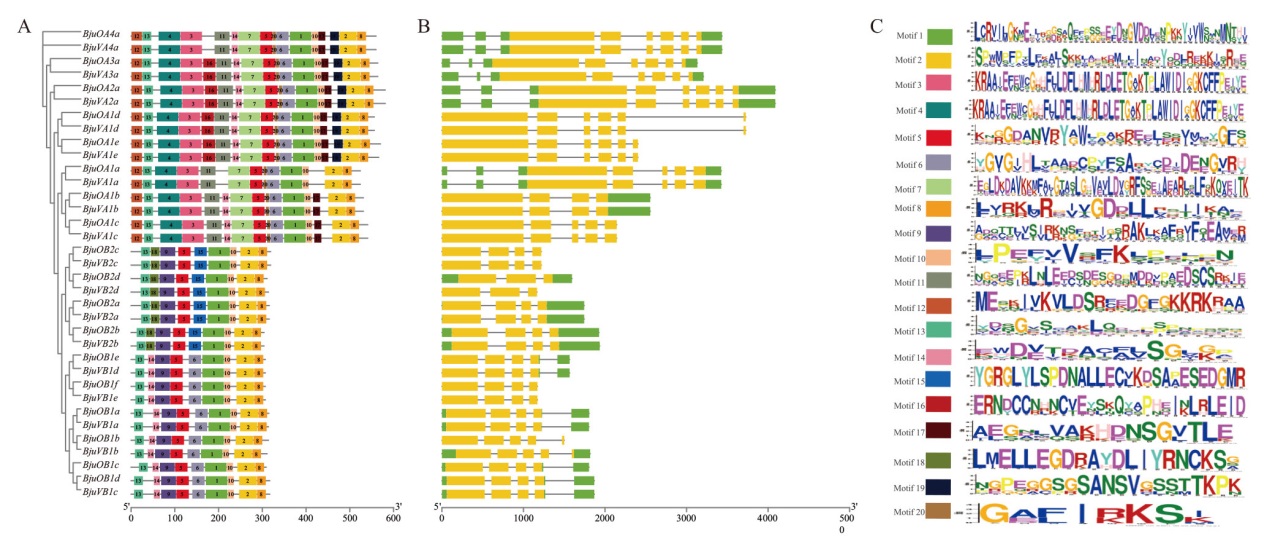
图1 BjuSRO基因结构分析 A:BjuSRO蛋白保守基序分析;B:BjuSRO基因结构分析;C:BjuSRO蛋白保守基序logo
Fig. 1 Structure analysis of BjuSRO gene A: Conserved motif analysis of BjuSRO protein. B: Structure analysis of BjuSRO gene. C: Conserved motif logo of BjuSRO protein

图2 BjuSRO蛋白结构域分析 A:SRO蛋白分类示意图;B:代表性BjuSRO蛋白3D结构图;C:BjuSRO蛋白PARP结构域序列比对;D:A类BjuSRO蛋白WWE结构域序列比对; E:BjuSRO蛋白SRT结构域序列比对
Fig. 2 Domain analysis of BjuSRO protein A: Schematic diagram of SRO protein classification. B: 3D structure diagram of representative BjuSRO protein. C: PARP domain sequence alignment of BjuSRO protein. D: WWE domain sequence alignment of class A BjuSRO protein. E: SRT domain sequence alignment of BjuSRO protein
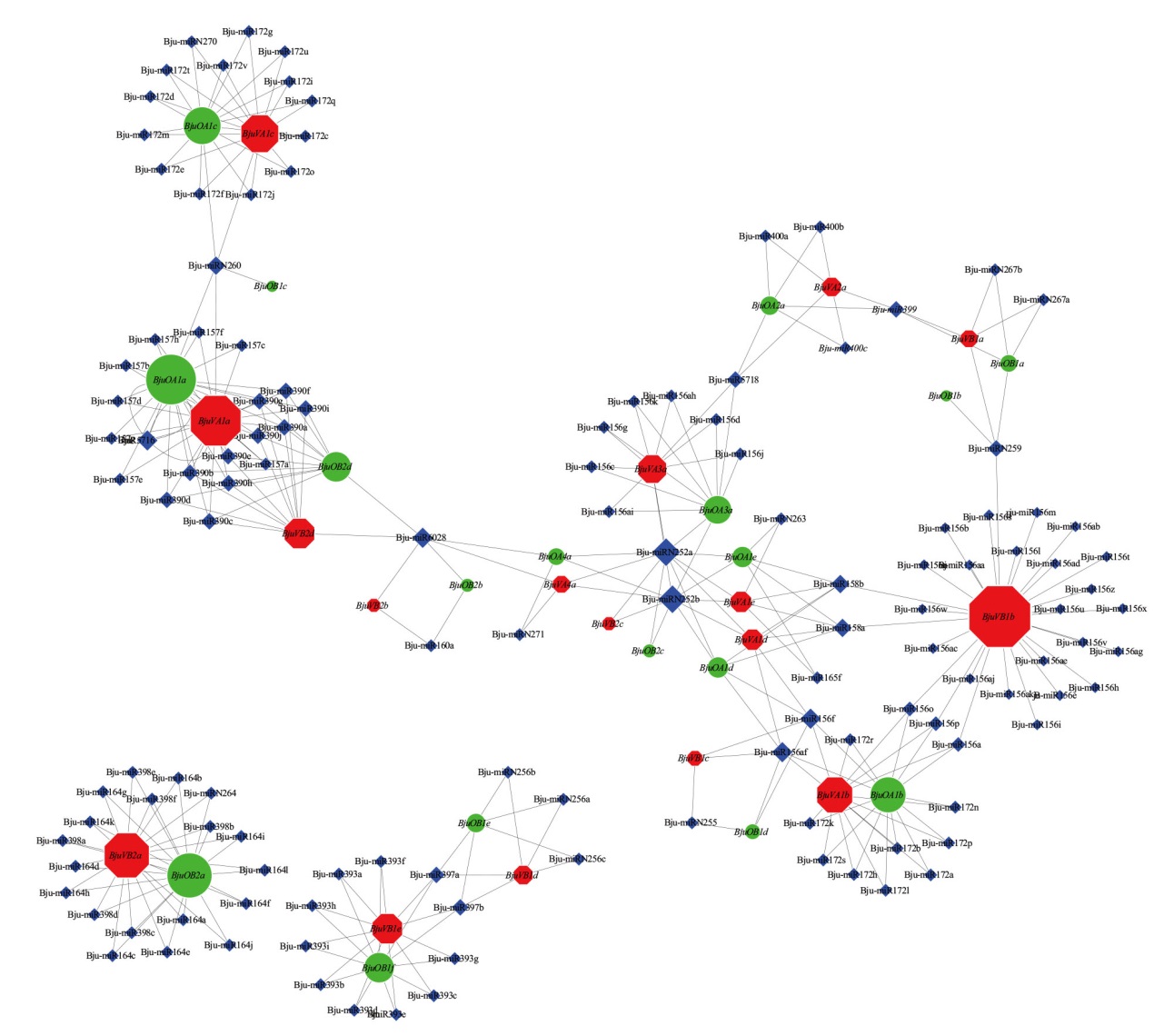
图4 Bju-miRNA与BjuSRO基因互作网络图 菱形表示Bju-miRNA;圆形表示油用芥菜BjuOSRO基因;八边形表示菜用芥菜BjuVSRO基因
Fig. 4 Network diagram of interaction between Bju-miRNA and BjuSRO gene Diamonds indicate Bju-miRNAs; circles indicate BjuOSRO gene of oilseed mustard; octagon indicate BjuVSRO gene of vegetable mustard
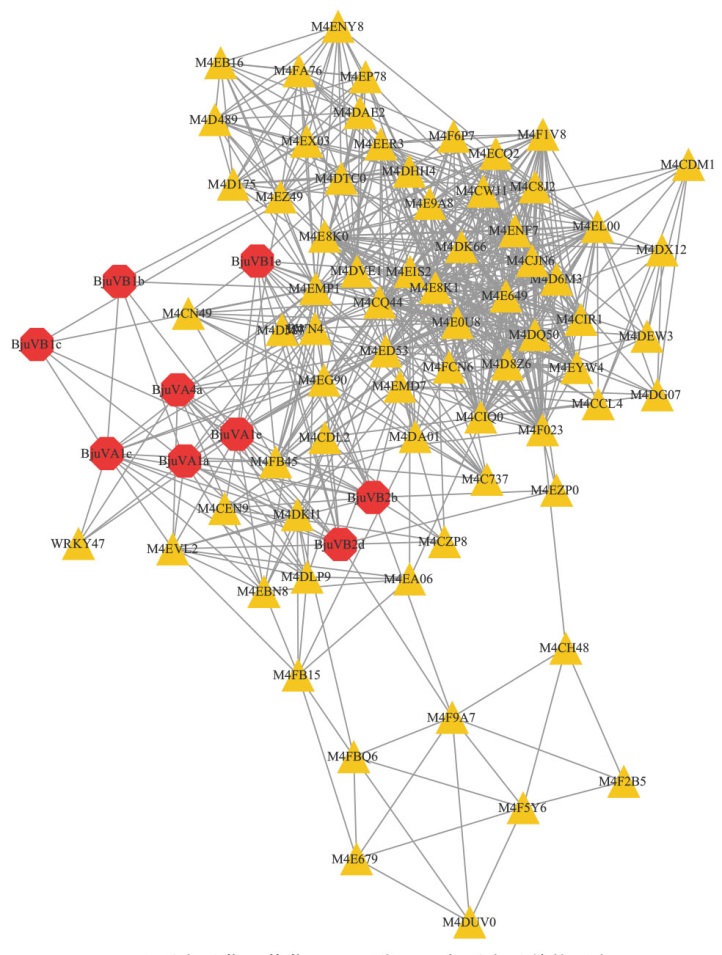
图6 BjuRSO蛋白互作分析网络图 八边形表示菜用芥菜SRO蛋白;三角形表示其他蛋白
Fig. 6 Network diagram of BjuRSO protein interaction analysis Octagon indicate SRO proteins of vegetable mustard; triangle indicate other proteins
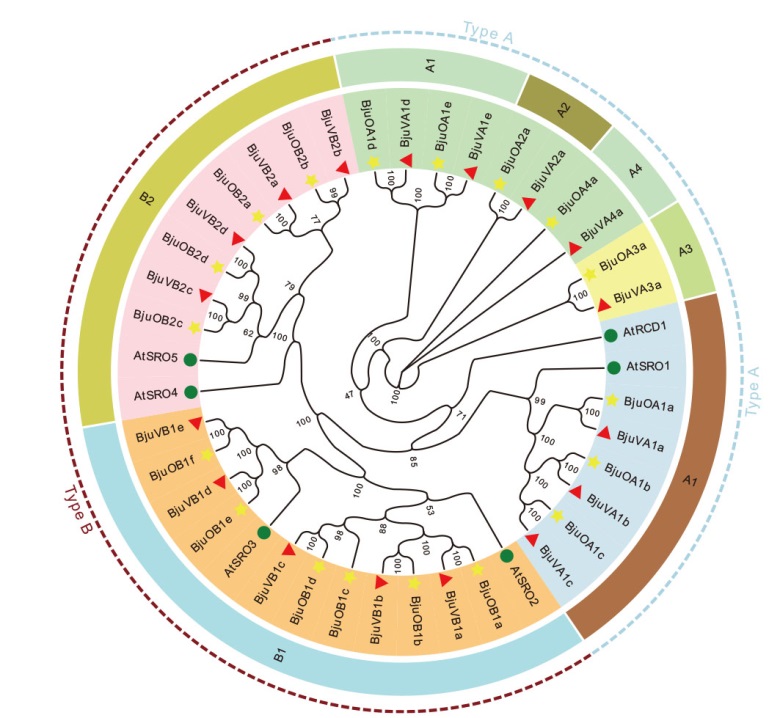
图7 SRO基因家族进化树分析 圆形表示拟南芥SRO蛋白;五角形表示油用芥菜SRO蛋白;三角形表示菜用芥菜SRO蛋白
Fig. 7 Phylogenetic tree analysis of SRO gene family Circle indicates SRO proteins of Arabidopsis; pentagon indicate SRO proteins of oilseed mustard; triangle indicate SRO proteins of vegetable mustard
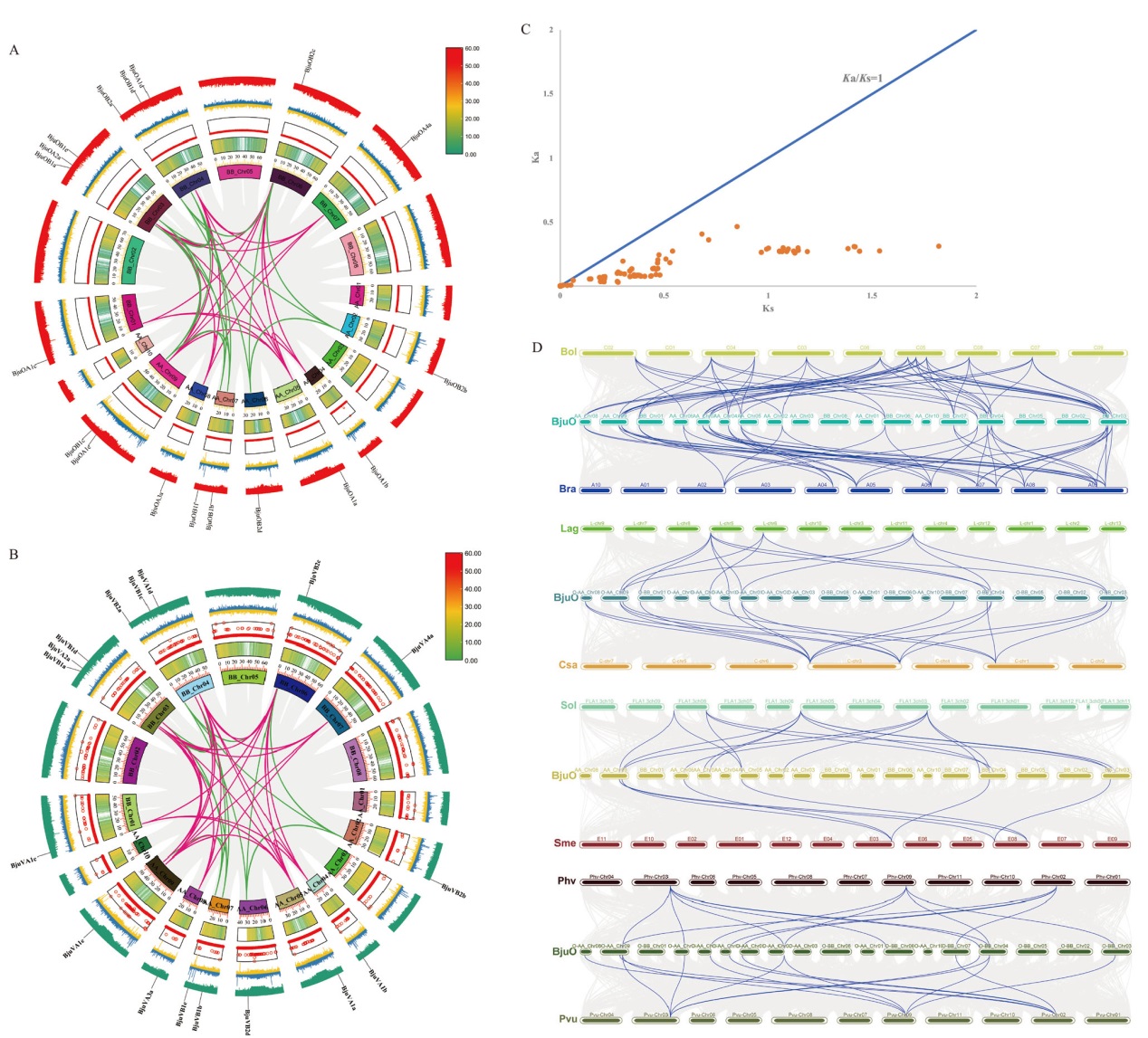
图8 SRO基因共线性分析 A:油用芥菜BjuOSRO基因共线性分析;B:菜用芥菜BjuVSRO基因共线性分析;图A和图B中的红线标记A类SRO基因,绿线标记B类SRO基因;C:芥菜SRO直系同源基因对Ka/Ks分析;D:油用芥菜与多物种SRO基因共线性分析(Bol:甘蓝;Bra:大白菜;Lag:丝瓜;Csa:黄瓜;Sol:番茄;Sme:茄子;Pvu:菜豆;Phv:豇豆)
Fig. 8 Collinearity analysis of SRO gene A: Collinear analysis of BjuOSRO gene in oil mustard. B: Collinear analysis of BjuVSRO gene in vegetable mustard. Red line marks class A SRO gene, and green line marks class B SRO gene in Fig. A and B. C: Ka/Ks analysis of SRO homologous genes in mustard. D: Collinearity analysis of SRO gene between oil mustard and multi species(Bol: Cabbage; Bra: Chinese cabbage; Lag: loofah; Csa: cucumber; Sol: tomato; Sme: eggplant; Pvu: kidney beans; Phv: cowpea)
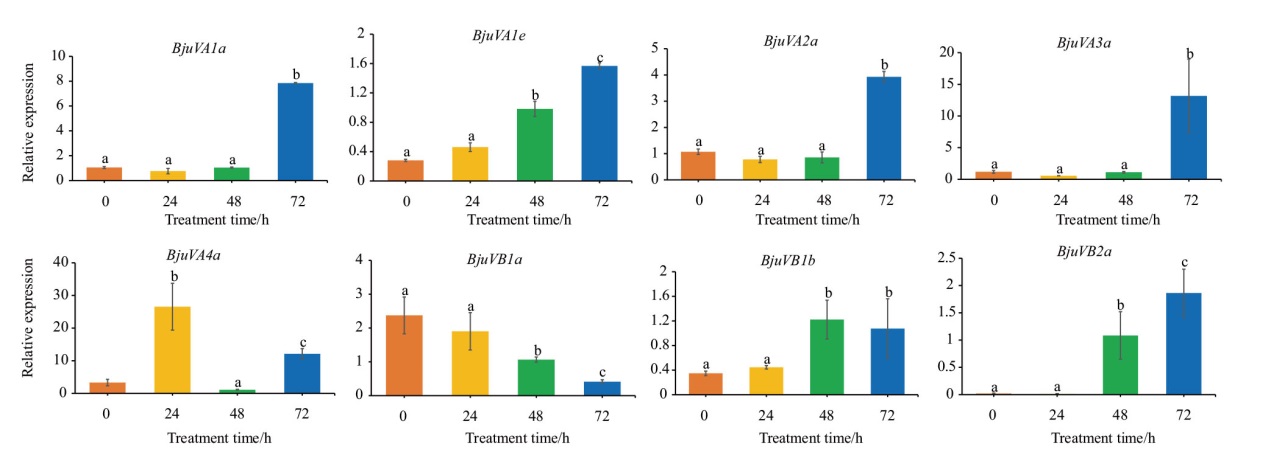
图9 不同时间盐胁迫下芥菜根系BjuSRO基因的相对表达量 不同小写字母表示差异显著(P < 0.05)
Fig. 9 Relative expressions of BjuSRO gene in mustard root system under salt stress at different times Different lowercase letters indicate significant differences(P<0.05)
| [1] | Zhang HM, Zhu JH, Gong ZZ, et al. Abiotic stress responses in plants[J]. Nat Rev Genet, 2022, 23(2): 104-119. |
| [2] |
Yang J, Ji LX, Liu S, et al. The CaM1-associated CCaMK-MKK1/6 cascade positively affects lateral root growth via auxin signaling under salt stress in rice[J]. J Exp Bot, 2021, 72(18): 6611-6627.
doi: 10.1093/jxb/erab287 pmid: 34129028 |
| [3] | Waadt R, Seller CA, Hsu PK, et al. Plant hormone regulation of abiotic stress responses[J]. Nat Rev Mol Cell Biol, 2022, 23(10): 680-694. |
| [4] | Chen H, Bullock DA Jr, Alonso JM, et al. To fight or to grow: the balancing role of ethylene in plant abiotic stress responses[J]. Plants, 2021, 11(1): 33. |
| [5] |
Chini A, Monte I, Fernández-Barbero G, et al. A small molecule antagonizes jasmonic acid perception and auxin responses in vascular and nonvascular plants[J]. Plant Physiol, 2021, 187(3): 1399-1413.
doi: 10.1093/plphys/kiab369 pmid: 34618088 |
| [6] | Zhang LC, He GH, Li YP, et al. PIL transcription factors directly interact with SPLs and repress tillering/branching in plants[J]. New Phytol, 2022, 233(3): 1414-1425. |
| [7] | Cheng ZY, Luan YT, Meng JS, et al. WRKY transcription factor response to high-temperature stress[J]. Plants, 2021, 10(10): 2211. |
| [8] | Liu J, Meng QL, Xiang HT, et al. Genome-wide analysis of Dof transcription factors and their response to cold stress in rice(Oryza sativa L.)[J]. BMC Genomics, 2021, 22(1): 800. |
| [9] |
Zhang HC, Shao SP, Zeng Y, et al. Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock[J]. Nat Cell Biol, 2022, 24(3): 340-352.
doi: 10.1038/s41556-022-00846-7 pmid: 35256776 |
| [10] | Verma S, Negi NP, Pareek S, et al. Auxin response factors in plant adaptation to drought and salinity stress[J]. Physiol Plant, 2022, 174(3): e13714. |
| [11] |
Alshareef NO, Otterbach SL, Allu AD, et al. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis[J]. Sci Rep, 2022, 12(1): 11264.
doi: 10.1038/s41598-022-14429-x pmid: 35787631 |
| [12] |
Jaspers P, Overmyer K, Wrzaczek M, et al. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants[J]. BMC Genomics, 2010, 11: 170.
doi: 10.1186/1471-2164-11-170 pmid: 20226034 |
| [13] | Jaspers P, Blomster T, Brosché M, et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors[J]. Plant J, 2009, 60(2): 268-279. |
| [14] |
Webb C, Upadhyay A, Giuntini F, et al. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway[J]. Biochemistry, 2011, 50(16): 3300-3309.
doi: 10.1021/bi2001888 pmid: 21417403 |
| [15] | Carter AA, Ramsey KM, Hatem CL, et al. Structural features of the Notch ankyrin domain-Deltex WWE2 domain heterodimer determined by NMR spectroscopy and functional implications[J]. Structure, 2023, 31(5): 584-594.e5. |
| [16] | Kim H, Aliar K, Tharmapalan P, et al. Differential DNA damage repair and PARP inhibitor vulnerability of the mammary epithelial lineages[J]. Cell Rep, 2023, 42(11): 113382. |
| [17] | Padella A, Ghelli Luserna Di Rorà A, Marconi G, et al. Targeting PARP proteins in acute leukemia: DNA damage response inhibition and therapeutic strategies[J]. J Hematol Oncol, 2022, 15(1): 10. |
| [18] | Maru B, Messikommer A, Huang LH, et al. PARP-1 improves leukemia outcomes by inducing parthanatos during chemotherapy[J]. Cell Rep Med, 2023, 4(9): 101191. |
| [19] |
Kong L, Feng BM, Yan Y, et al. Noncanonical mono(ADP-ribosyl)ation of zinc finger SZF proteins counteracts ubiquitination for protein homeostasis in plant immunity[J]. Mol Cell, 2021, 81(22): 4591-4604.e8.
doi: 10.1016/j.molcel.2021.09.006 pmid: 34592134 |
| [20] |
Vainonen JP, Jaspers P, Wrzaczek M, et al. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana[J]. Biochem J, 2012, 442(3): 573-581.
doi: 10.1042/BJ20111739 pmid: 22150398 |
| [21] |
Ahlfors R, Lång S, Overmyer K, et al. Arabidopsis radical-induced cell death1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses[J]. Plant Cell, 2004, 16(7): 1925-1937.
doi: 10.1105/tpc.021832 pmid: 15208394 |
| [22] | Xu W, Zhu WW, Yang L, et al. SMALL REPRODUCTIVE ORGANS, a SUPERMAN-like transcription factor, regulates stamen and pistil growth in rice[J]. New Phytol, 2022, 233(4): 1701-1718. |
| [23] |
You J, Zong W, Du H, et al. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors[J]. Plant Mol Biol, 2014, 84(6): 693-705.
doi: 10.1007/s11103-013-0163-8 pmid: 24337801 |
| [24] | Li HH, Li R, Qu FJ, et al. Identification of the SRO gene family in apples(Malus × domestica)with a functional characterization of MdRCD1[J]. Tree Genet Genomes, 2017, 13(5): 94. |
| [25] | Yuan BQ, Chen MH, Li SS. Isolation and identification of Ipomoea cairica(L.)sweet gene IcSRO1 encoding a SIMILAR TO RCD-ONE protein, which improves salt and drought tolerance in transgenic Arabidopsis[J]. Int J Mol Sci, 2020, 21(3): 1017. |
| [26] | Du YJ, Roldan MVG, Haraghi A, et al. Spatially expressed WIP genes control Arabidopsis embryonic root development[J]. Nat Plants, 2022, 8(6): 635-645. |
| [27] | Li XY, Xu YJ, Liu F, et al. Maize similar to RCD1 gene induced by salt enhances Arabidopsis thaliana abiotic stress resistance[J]. Biochem Biophys Res Commun, 2018, 503(4): 2625-2632. |
| [28] | Sanlier N, Guler SM. The benefits of Brassica vegetables on human health[J]. Journal of Human Health Research, 2018, 1(1): 104. |
| [29] | Yang JH, Wang J, Li ZP, et al. Genomic signatures of vegetable and oilseed allopolyploid Brassica juncea and genetic loci controlling the accumulation of glucosinolates[J]. Plant Biotechnol J, 2021, 19(12): 2619-2628. |
| [30] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [31] | Islam S, Shah SH, Corpas FJ, et al. Plant growth regulators mediated mitigation of salt-induced toxicities in mustard(Brassica juncea L.)by modifying the inherent defense system[J]. Plant Physiol Biochem, 2023, 196: 1002-1018. |
| [32] | Dixit S, Jangid VK, Grover A. Evaluation of suitable reference genes in Brassica juncea and its wild relative Camelina sativa for RT-qPCR analysis under various stress conditions[J]. PLoS One, 2019, 14(9): e0222530. |
| [33] |
Dizengremel P, Le Thiec D, Bagard M, et al. Ozone risk assessment for plants: central role of metabolism-dependent changes in reducing power[J]. Environ Pollut, 2008, 156(1): 11-15.
doi: 10.1016/j.envpol.2007.12.024 pmid: 18243452 |
| [34] | Li BZ, Zhao X, Zhao XL, et al. Structure and function analysis of Arabidopsis thaliana SRO protein family[J]. Yi Chuan, 2013, 35(10): 1189-1197. |
| [35] | Qiao YL, Gao XQ, Liu ZC, et al. Genome-wide identification and analysis of SRO gene family in Chinese cabbage(Brassica rapa L.)[J]. Plants, 2020, 9(9): 1235. |
| [36] | Vainonen JP, Shapiguzov A, Krasensky-Wrzaczek J, et al. Arabidopsis poly(ADP-ribose)-binding protein RCD1 interacts with photoregulatory protein kinases in nuclear bodies[J]. bioRxiv, 2020. https://doi.org/10.1101/2020.07.02.184937. |
| [37] | Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements[J]. Plant Sci, 2014, 217/218: 109-119. |
| [38] |
Lu TX, Rothenberg ME. MicroRNA[J]. J Allergy Clin Immunol, 2018, 141(4): 1202-1207.
doi: S0091-6749(17)31593-2 pmid: 29074454 |
| [39] |
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods[J]. J Cell Physiol, 2019, 234(5): 5451-5465.
doi: 10.1002/jcp.27486 pmid: 30471116 |
| [40] | Li N, Xu RQ, Wang BK, et al. Genome-wide identification and evolutionary analysis of the SRO gene family in tomato[J]. Front Genet, 2021, 12: 753638. |
| [41] |
Zhang L, Zhou DB, Hu HG, et al. Genome-wide characterization of a SRO gene family involved in response to biotic and abiotic stresses in banana(Musa spp.)[J]. BMC Plant Biol, 2019, 19(1): 211.
doi: 10.1186/s12870-019-1807-x pmid: 31113386 |
| [42] | Jiang WQ, Geng YP, Liu YK, et al. Genome-wide identification and characterization of SRO gene family in wheat: molecular evolution and expression profiles during different stresses[J]. Plant Physiol Biochem, 2020, 154: 590-611. |
| [43] | Qin LM, Sun L, Wei L, et al. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress[J]. Plant J, 2021, 105(4): 1010-1025. |
| [44] | Liu AL, Wei MY, Zhou Y, et al. Comprehensive analysis of SRO gene family in Sesamum indicum(L.)reveals its association with abiotic stress responses[J]. Int J Mol Sci, 2021, 22(23): 13048. |
| [45] |
Song XW, Li Y, Cao XF, et al. MicroRNAs and their regulatory roles in plant-environment interactions[J]. Annu Rev Plant Biol, 2019, 70: 489-525.
doi: 10.1146/annurev-arplant-050718-100334 pmid: 30848930 |
| [46] | Singh I, Smita S, Mishra DC, et al. Abiotic stress responsive miRNA-target network and related markers(SNP, SSR)in Brassica juncea[J]. Front Plant Sci, 2017, 8: 1943. |
| [47] | Wang YT, Wang RQ, Yu Y, et al. Genome-wide analysis of SIMILAR TO RCD ONE(SRO)family revealed their roles in abiotic stress in poplar[J]. Int J Mol Sci, 2023, 24(4): 4146. |
| [1] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| [2] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| [3] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [4] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [5] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [6] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| [7] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [8] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [9] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [10] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [11] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [12] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [13] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [14] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [15] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||