








生物技术通报 ›› 2025, Vol. 41 ›› Issue (11): 282-292.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0389
• 研究报告 • 上一篇
收稿日期:2025-04-13
出版日期:2025-11-26
发布日期:2025-12-09
通讯作者:
李瑞民,男,博士,副教授,研究方向 :柑橘抗病育种;E-mail: liruimin@gnnu.edu.cn作者简介:胡衍铵,男,硕士研究生,研究方向 :柑橘黄龙病菌致病机制解析;E-mail: huyanan@gnnu.edu.cn
基金资助:
HU Yan-an( ), DAI Xin-lyu, ZHONG Jiao-yan, LI Rui-min(
), DAI Xin-lyu, ZHONG Jiao-yan, LI Rui-min( ), HUANG Gui-yan(
), HUANG Gui-yan( )
)
Received:2025-04-13
Published:2025-11-26
Online:2025-12-09
摘要:
目的 耐病基因的发掘是培育耐黄龙病(Huanglongbing, HLB)柑橘材料的重要基础,为解析柑橘与黄龙病菌互作的分子机制,筛选柑橘属(Citrus spp.)及近缘种植物中耐黄龙病的关键基因。 方法 基于比较基因组学方法,系统鉴定了耐病材料中的关键基因,并利用特异性基因簇鉴定及共有直系同源群的分析,结合gene ontology(GO)富集分析,转录组数据分析以及蛋白质-蛋白质互作网络分析,鉴定耐病材料中的核心差异表达基因。 结果 黄龙病菌诱导的差异表达的防御相关基因中多个HSP70基因表达模式受黄龙病菌侵染的显著调控,而过表达HSP70显著提高了柑橘对细菌性病害的抗病性。 结论 耐病材料中防御相关基因HSP70正调控柑橘对病原菌的抗病性。
胡衍铵, 代鑫吕, 钟娇艳, 李瑞民, 黄桂艳. 基于比较基因组学的柑橘耐黄龙病关键基因鉴定与功能解析[J]. 生物技术通报, 2025, 41(11): 282-292.
HU Yan-an, DAI Xin-lyu, ZHONG Jiao-yan, LI Rui-min, HUANG Gui-yan. Comparative Genomics-based Identification and Functional Characterization of Key Genes Resisting Huanglongbing in Citrus[J]. Biotechnology Bulletin, 2025, 41(11): 282-292.

图1 11个物种基因家族聚类分析Proteins代表各物种非冗余蛋白序列,Clusters代表各物种直系同源群,Singletons代表各物种单拷贝基因簇
Fig. 1 Clustering analysis of gene families across 11 speciesProteins indicate the non-redundant protein sequences of each species. Clusters indicate the orthogroups (orthologous gene clusters) across species. Singletons indicate the single-copy gene clusters specific to each species
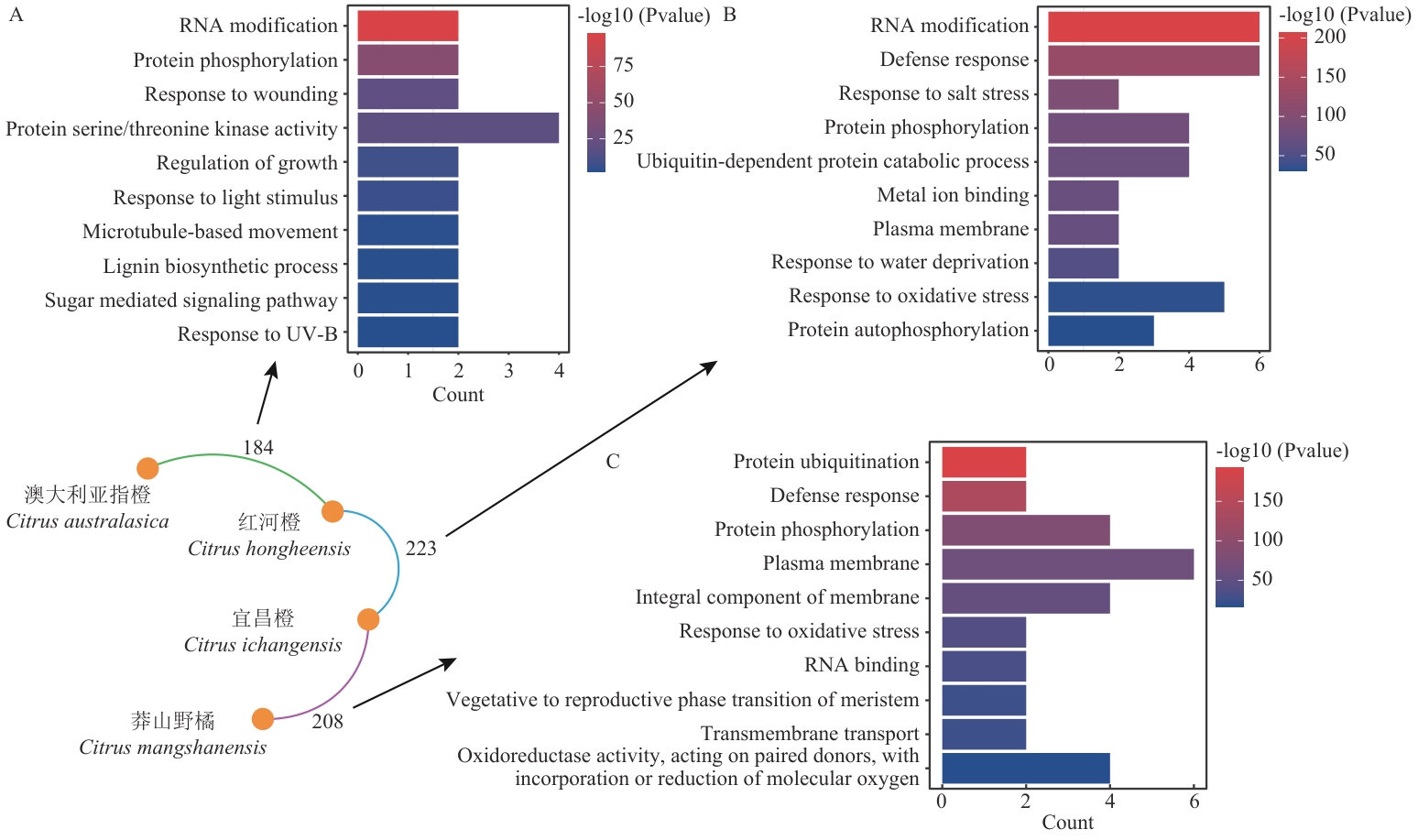
图3 耐黄龙病材料共有直系同源群蛋白GO富集分析A:澳大利亚指橙和红河橙;B:宜昌橙和红河橙;C:莽山野橘和宜昌橙
Fig. 3 GO enrichment analysis of proteins in common orthogroups from HLB-resistant speciesA: Citrus australasica and C. hongheensis; B: C. ichangensis and C. hongheensis; C: C. mangshanensis and C. ichangensis
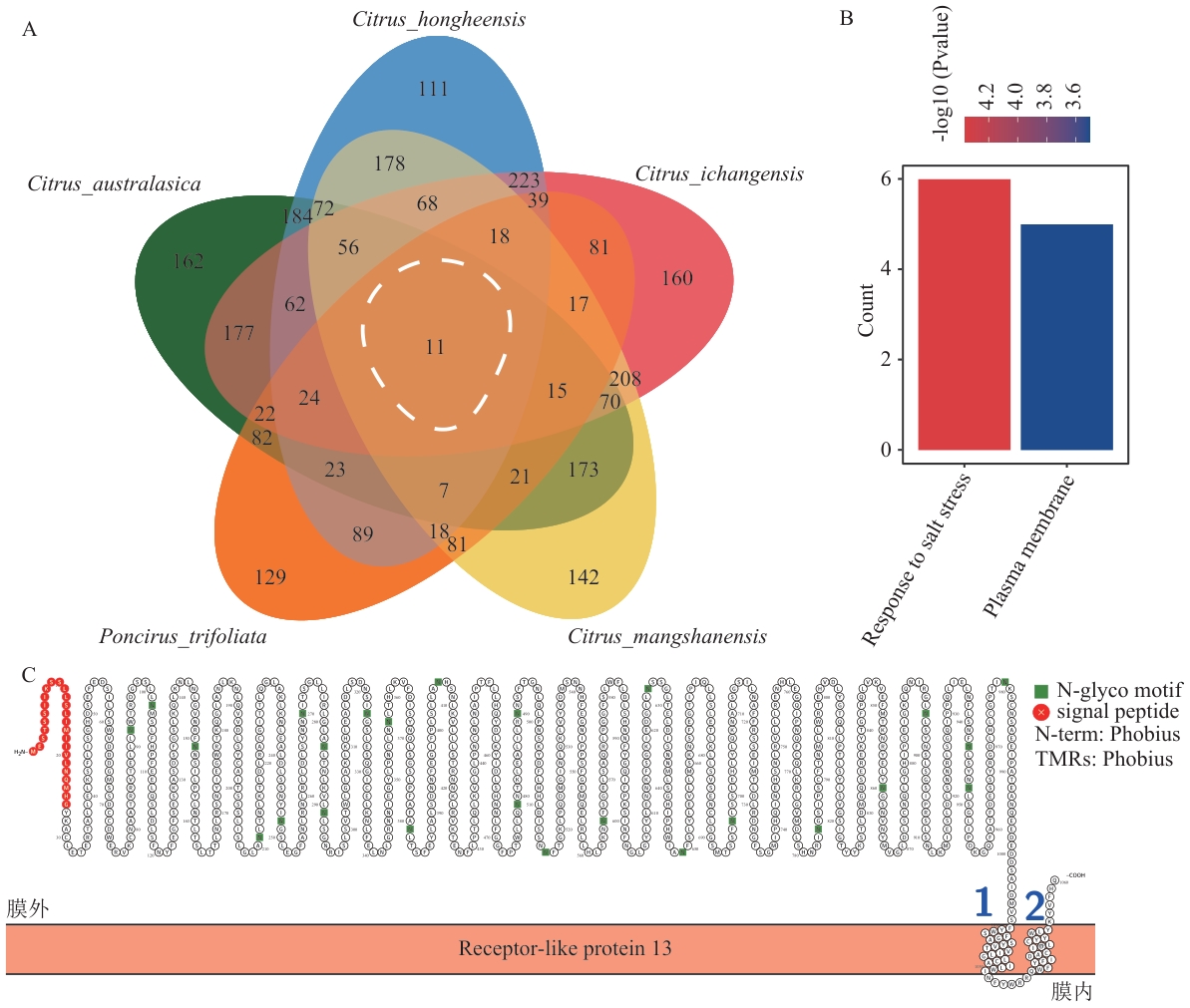
图4 五个耐黄龙病物种共有直系同源群分析A:共有直系同源群分析;B:GO富集分析;C:跨膜结构域分析
Fig. 4 Common orthogroups analysis from five HLB-resistant speciesA: Common orthogroups analysis; B: GO enrichment analysis; C: transmembrane domain analysis
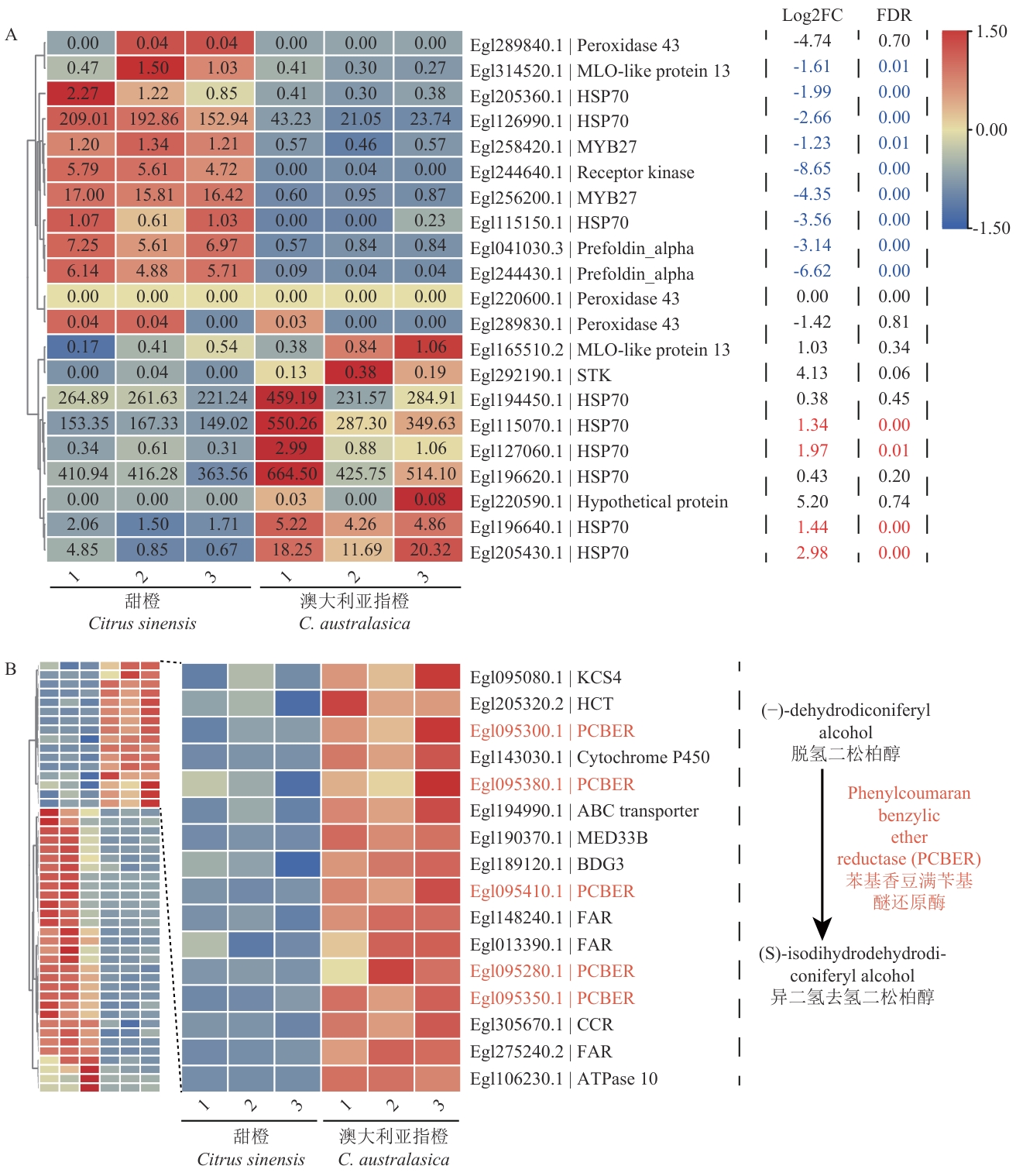
图7 澳大利亚指橙基因组中扩张基因家族相关基因的表达模式A:“防御反应”相关基因; B:“苯丙烷代谢过程”相关基因
Fig. 7 Expression patterns of genes related to the expanded gene family in C. australasicaA: “Defense response” related genes; B: “phenylpropanoid metabolic process” related genes
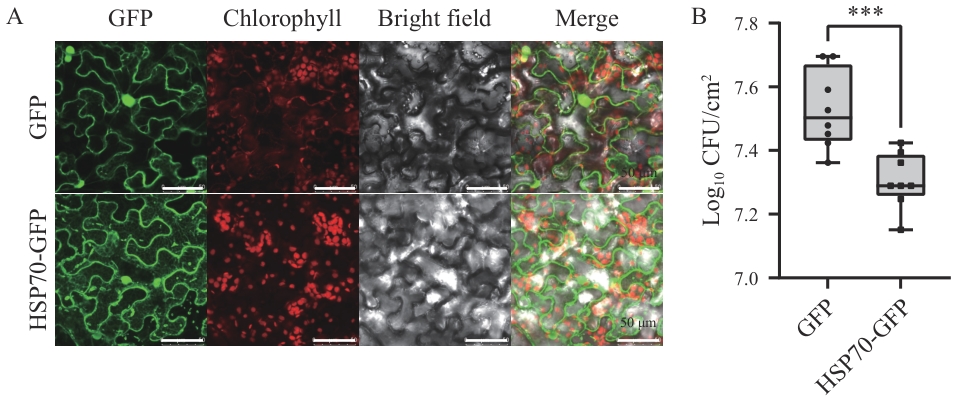
图9 HSP70的亚细胞定位及抗病性分析A:HSP70的亚细胞定位,标尺=50 μm;B:过表达HSP70对溃疡病的抗病性分析。以t-test进行统计学显著性分析,***表示P<0.001
Fig. 9 Subcellular localization and disease resistance analysis of HSP70A: Subcellular localization of HSP70, scale bar=50 μm. B: Analysis of the disease resistance of overexpressed HSP70 to citrus canker. Used t-test for statistical significance analysis, *** denotes P<0.001
| [1] | Wang N. The Citrus huanglongbing crisis and potential solutions [J]. Mol Plant, 2019, 12(5): 607-609. |
| [2] | Gottwald TR. Current epidemiological understanding of Citrus huanglongbing [J]. Annu Rev Phytopathol, 2010, 48: 119-139. |
| [3] | Hu YN, Lu NN, Bao KQ, et al. Swords and shields: the war between Candidatus Liberibacter asiaticus and Citrus [J]. Front Plant Sci, 2025, 15: 1518880. |
| [4] | Waengwan P, Laosatit K, Lin Y, et al. A cluster of Peronospora parasitica 13-like (NBS-LRR) genes is associated with powdery mildew (Erysiphe polygoni) resistance in mungbean (Vigna radiata) [J]. Plants, 2024, 13(9): 1230. |
| [5] | Patial M, Navathe S, He XY, et al. Novel resistance loci for quantitative resistance to Septoria tritici blotch in Asian wheat (Triticum aestivum) via genome-wide association study [J]. BMC Plant Biol, 2024, 24(1): 846. |
| [6] | Luo YY, Liu ZY, Jin ZX, et al. Phased T2T genome assemblies facilitate the mining of disease-resistance genes in Vitis davidii [J]. Hortic Res, 2024, 12(2): uhae306. |
| [7] | Wu GA, Terol J, Ibanez V, et al. Genomics of the origin and evolution of Citrus [J]. Nature, 2018, 554(7692): 311-316. |
| [8] | Wang L, He F, Huang Y, et al. Genome of wild mandarin and domestication history of mandarin [J]. Mol Plant, 2018, 11(8): 1024-1037. |
| [9] | Wang FS, Wang SH, Wu YL, et al. Haplotype-resolved genome of a papeda provides insights into the geographical origin and evolution of citrus [J]. J Integr Plant Biol, 2025, 67(2): 276-293. |
| [10] | Huang Y, He JX, Xu YT, et al. Pangenome analysis provides insight into the evolution of the orange subfamily and a key gene for citric acid accumulation in citrus fruits [J]. Nat Genet, 2023, 55(11): 1964-1975. |
| [11] | Huang Y, Makkumrai W, Fu JL, et al. Genomic analysis provides insights into the origin and divergence of fruit flavor and flesh color of pummelo [J]. New Phytol, 2025, 245(1): 378-391. |
| [12] | Ramadugu C, Keremane ML, Halbert SE, et al. Long-term field evaluation reveals huanglongbing resistance in citrus relatives [J]. Plant Dis, 2016, 100(9): 1858-1869. |
| [13] | Alves MN, Lopes SA, Raiol-Junior LL, et al. Resistance to Candidatus liberibacter asiaticus, the huanglongbing associated bacterium, in sexually and/or graft-compatible citrus relatives [J]. Front Plant Sci, 2021, 11: 617664. |
| [14] | Weber KC, Mahmoud LM, Stanton D, et al. Insights into the mechanism of Huanglongbing tolerance in the Australian finger lime (Citrus australasica) [J]. Front Plant Sci, 2022, 13: 1019295. |
| [15] | Peng Z, Bredeson JV, Wu GA, et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus [J]. Plant J, 2020, 104(5): 1215-1232. |
| [16] | Nakandala U, Furtado A, Masouleh AK, et al. The genome of Citrus australasica reveals disease resistance and other species specific genes [J]. BMC Plant Biol, 2024, 24(1): 260. |
| [17] | Liu H, Wang X, Liu SJ, et al. Citrus pan-genome to breeding database (CPBD): a comprehensive genome database for Citrus breeding [J]. Mol Plant, 2022, 15(10): 1503-1505. |
| [18] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [19] | Garcia-Hernandez M, Berardini TZ, Chen GH, et al. TAIR: a resource for integrated Arabidopsis data [J]. Funct Integr Genomics, 2002, 2(6): 239-253. |
| [20] | Sun JH, Lu F, Luo YJ, et al. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes [J]. Nucleic Acids Res, 2023, 51(W1): W397-W403. |
| [21] | Cantalapiedra CP, Hernández-Plaza A, Letunic I, et al. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale [J]. Mol Biol Evol, 2021, 38(12): 5825-5829. |
| [22] | Bolger M, Schwacke R, Usadel B. MapMan visualization of RNA-seq data using Mercator4 functional annotations [J]. Methods Mol Biol, 2021, 2354: 195-212. |
| [23] | Schwacke R, Ponce-Soto GY, Krause K, et al. MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis [J]. Mol Plant, 2019, 12(6): 879-892. |
| [24] | Mu HY, Chen JZ, Huang WJ, et al. OmicShare tools: a zero-code interactive online platform for biological data analysis and visualization [J]. Imeta, 2024, 3(5): e228. |
| [25] | Supek F, Bošnjak M, Škunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms [J]. PLoS One, 2011, 6(7): e21800. |
| [26] | Chen SF, Zhou YQ, Chen YR, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor [J]. Bioinformatics, 2018, 34(17): i884-i890. |
| [27] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 [J]. Genome Biol, 2014, 15(12): 550. |
| [28] | Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest [J]. Nucleic Acids Res, 2023, 51(D1): D638-D646. |
| [29] | Wu HD, Hu Y, Fu SM, et al. Coordination of multiple regulation pathways contributes to the tolerance of a wild citrus species (citrus ichangensis ‘2586’) against Huanglongbing [J]. Physiol Mol Plant Pathol, 2020, 109: 101457. |
| [30] | 唐艳, 武晓晓, 娄兵海, 等. 一种快速鉴定柑橘黄龙病耐性种质材料的方法 [J]. 南方农业学报, 2019, 50(11): 2489-2495. |
| Tang Y, Wu XX, Lou BH, et al. A rapid identification method for germplasm materials resistant to citrus Huanglongbing disease [J]. J South Agric, 2019, 50(11): 2489-2495. | |
| [31] | Mou FJ, Li JQ, Gao YH, et al. Morphological characteristics, genetic diversity, and ethnobotanical uses of Citrus Hongheensis(Rutaceae), an endangered species [J]. Plant Biosyst Int J Deal Aspects Plant Biol, 2024, 158(3): 464-472. |
| [32] | Sohn KH, Lee SC, Jung HW, et al. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance [J]. Plant Mol Biol, 2006, 61(6): 897-915. |
| [33] | Zhao H, Jiang J, Li KL, et al. Populus simonii × Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses [J]. Tree Physiol, 2017, 37(6): 827-844. |
| [34] | Asano T, Hayashi N, Kobayashi M, et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance [J]. Plant J, 2012, 69(1): 26-36. |
| [35] | Tör M, Lotze MT, Holton N. Receptor-mediated signalling in plants: molecular patterns and programmes [J]. J Exp Bot, 2009, 60(13): 3645-3654. |
| [36] | DeFalco TA, Zipfel C. Molecular mechanisms of early plant pattern-triggered immune signaling [J]. Mol Cell, 2021, 81(17): 3449-3467. |
| [37] | Jacquemin J, Ammiraju JSS, Haberer G, et al. Fifteen million years of evolution in the Oryza genus shows extensive gene family expansion [J]. Mol Plant, 2014, 7(4): 642-656. |
| [38] | Zhang QJ, Zhu T, Xia EH, et al. Rapid diversification of five Oryza AA genomes associated with rice adaptation [J]. Proc Natl Acad Sci USA, 2014, 111(46): E4954-E4962. |
| [39] | Cintron NS, Toft D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway [J]. J Biol Chem, 2006, 281(36): 26235-26244. |
| [40] | Berka M, Kopecká R, Berková V, et al. Regulation of heat shock proteins 70 and their role in plant immunity [J]. J Exp Bot, 2022, 73(7): 1894-1909. |
| [41] | Kanzaki H, Saitoh H, Ito A, et al. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana [J]. Mol Plant Pathol, 2003, 4(5): 383-391. |
| [42] | Liu JZ, Whitham SA. Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance [J]. Plant J, 2013, 74(1): 110-121. |
| [43] | Brugière N, Zhang WJ, Xu QZ, et al. Overexpression of RING domain E3 ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis [J]. Plant Physiol, 2017, 175(3): 1350-1369. |
| [44] | Marino D, Froidure S, Canonne J, et al. Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence [J]. Nat Commun, 2013, 4: 1476. |
| [45] | Guo XR, Su J, Xue H, et al. Genome-wide identification and expression analyses of ABSCISIC ACID-INSENSITIVE 5 (ABI5) genes in Citrus sinensis reveal CsABI5-5 confers dual resistance to Huanglongbing and citrus canker [J]. Int J Biol Macromol, 2025, 306(Pt 4): 141611. |
| [1] | 苗昊翔, 张颖, 郭世鹏, 张健. 一株高产γ-氨基丁酸短乳杆菌TCCC13007全基因组测序及比较基因组分析[J]. 生物技术通报, 2025, 41(11): 166-176. |
| [2] | 雷美玲, 饶文华, 胡进锋, 岳琪, 吴祖建, 范国成. 黄龙病发病芦柑根际土壤细菌群落组成与多样性特征[J]. 生物技术通报, 2024, 40(2): 266-276. |
| [3] | 张泽颖, 范清锋, 邓云峰, 韦廷舟, 周正富, 周建, 王劲, 江世杰. 一株高产脂肪酶菌株WCO-9全基因组测序及比较基因组分析[J]. 生物技术通报, 2022, 38(10): 216-225. |
| [4] | 张钰, 海萨·艾也力汗, 热比古丽·沙吾提, 时春明, 张人铭. 白斑狗鱼早期发育阶段的高温耐受性分析[J]. 生物技术通报, 2021, 37(5): 76-83. |
| [5] | 樊晓猛, 戚继. 基于比较基因组学方法揭示十字花科古老杂交事件[J]. 生物技术通报, 2018, 34(7): 126-137. |
| [6] | 赵志文, 李艳娇, 户勋, 范晓静, 卓涛, 邹华松. 用于植物病原细菌标记的pBB-GFP载体构建及应用[J]. 生物技术通报, 2018, 34(3): 136-141. |
| [7] | 王龑, 刘阳, 刘伊宁. 产毒真菌基因组研究进展[J]. 生物技术通报, 2015, 31(2): 26-34. |
| [8] | 高雪;. 柑橘钙结合蛋白基因的克隆及其在果皮褐变中的表达分析[J]. , 2011, 0(06): 53-57. |
| [9] | 高雪;. 柑橘膨胀素相似基因的克隆及在采后胁迫条件下的表达[J]. , 2010, 0(09): 87-92. |
| [10] | 高雪;. 一种适合从柑橘果皮提取总RNA的方法[J]. , 2010, 0(07): 134-136. |
| [11] | 艾华水;胡小芬;周利华;. 猪Mitf基因的比较基因组学和进化分析[J]. , 2010, 0(07): 161-167. |
| [12] | 施维属;潘腾飞;钟凤林;潘东明;李开拓;王江波;郭志雄;刘天亮;. 柑橘基因组DNA快速提取及ISSR-PCR扩增体系优化[J]. , 2009, 0(10): 109-113. |
| [13] | 廖惠红;白先进;李杨瑞;陈传武;杨丽涛;朱建华;徐宁;. 基于23S/5S rDNA序列的柑橘黄龙病病原菌亚洲种系统发育研究[J]. , 2009, 0(05): 88-91. |
| [14] | 黎裕;王天宇. 玉米比较基因组学研究进展[J]. , 2004, 0(01): 23-26. |
| [15] | 黎裕;王天宇;贾继增. 植物比较基因组学的研究进展[J]. , 2000, 0(05): 11-14. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||