








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 150-160.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0598
杨娟( ), 冯慧, 吉乃喆, 孙丽萍, 王赟, 张佳楠, 赵世伟(
), 冯慧, 吉乃喆, 孙丽萍, 王赟, 张佳楠, 赵世伟( )
)
收稿日期:2025-06-10
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
赵世伟,男,博士,教授级高工,研究方向 :园林植物育种;E-mail: 2668587780@qq.com作者简介:杨娟,女,博士,工程师,研究方向 :园林植物育种;E-mail: yangjuanyl@126.com
基金资助:
YANG Juan( ), FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei(
), FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei( )
)
Received:2025-06-10
Published:2026-01-26
Online:2026-02-04
摘要:
目的 AP2/ERF(APETALA2/ethylene-responsive factor)转录因子家族在调控植物生长发育、类黄酮合成和响应逆境胁迫等方面发挥重要作用。克隆RcERF4和RcRAP2-12基因并分析其功能,为揭示AP2/ERF转录因子调控月季花色中的作用奠定基础。 方法 首先,通过RT-PCR技术从月季‘夏洛特夫人’中克隆RcERF4和RcRAP2-12,并进行生物信息学、表达模式、亚细胞定位的分析及检测;进一步,采用农杆菌介导的叶盘转化法创制转RcERF4和RcRAP2-12基因烟草阳性植株,观察转基因烟草表型;最后,利用酵母双杂交和双分子荧光互补试验验证RcERF4和RcRAP2-12蛋白之间的相互作用关系。 结果 RcERF4的ORF(open reading frame)区为795 bp,编码264个氨基酸,RcRAP2-12的ORF区为1 179 bp,编码392个氨基酸;RcERF4和RcRAP2-12蛋白均含有1个AP2结构域,属于ERF亚族,与玫瑰、野草莓进化关系较近,蛋白同源性较高;RcERF4和RcRAP2-12主要在月季花瓣、雄蕊及子房中表达,属于花特异性表达基因,RcERF4在‘夏洛特夫人’花发育初期表达量较高,RcRAP2-12在整个花发育时期呈现出上调表达趋势;RcERF4和RcRAP2-12蛋白均定位于细胞核;在烟草中过量表达RcRAP2-12会提高烟草花冠中花青素含量和NtANS、NtUFGT、NtAN2的表达;RcERF4和RcRAP2-12存在相互作用关系。 结论 RcERF4和RcRAP2-12为细胞核定位的ERF类转录因子,RcRAP2-12可能在月季花色形成与调控中具有潜在功能。
杨娟, 冯慧, 吉乃喆, 孙丽萍, 王赟, 张佳楠, 赵世伟. 月季AP2/ERF转录因子RcERF4和RcRAP2-12的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 150-160.
YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose[J]. Biotechnology Bulletin, 2026, 42(1): 150-160.

图1 ‘夏洛特夫人’的不同花发育时期S1:萼片包裹紧实,花瓣未着色;S2:萼片张开,花瓣开始着色;S3:萼片完全张开,花瓣完全着色;S4:花瓣完全盛开。下同
Fig. 1 Different flower development stages of 'Lady of Shalott'S1: Sepals tightly wrapped, petals not colored. S2: Sepals open, petals begin to color. S3: Sepals fully open, petals fully colored. S4: Petals fully bloomed. The same below
| 用途 Application | 引物名称Primer name | 引物序列Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene cloning | RcERF4-F1 | ATGGCACCGAGAACCGAGA |
| RcERF4-R1 | TCAGGCGAGTTCCGCCGGA | |
| RcRAP2-12-F1 | ATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R1 | GAAAACTCCCCCAGACATTG | |
实时荧光定量PCR Quantitative real-time PCR | RcERF4-F2 | ACCACGGCTAAACCTAACGG |
| RcERF4-R2 | ATTCACATTCTCCGACGGCA | |
| RcRAP2-12-F2 | CGACTTGGTCTGGCCTGATT | |
| RcRAP2-12-R2 | TGCACTCCACGGATTTCACA | |
亚细胞定位和遗传转化烟草 Subcellular localization and genetic transformation of tobacco | RcERF4-F3 | aagtccggagctagctctagaATGGCACCGAGAACCGAGA |
| RcERF4-R3 | gcccttgctcaccatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F3 | aagtccggagctagctctagaATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R3 | gcccttgctcaccatggatccGAAAACTCCCCCAGACATTGAA | |
酵母双杂交 Yeast two-hybrid | RcERF4-F4 | gccatggaggccagtgaattcATGGCACCGAGAACCGAGA |
| RcERF4-R4 | cagctcgagctcgatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F4 | tcagaggaggacctgcatatgATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R4 | ctagttatgcggccgctgcagGAAAACTCCCCCAGACATTGAA | |
双分子荧光互补 Bimolecular fluorescence complementation | RcERF4-F5 | tggcgcgccactagtggatccATGGCACCGAGAACCGAGA |
| RcERF4-R5 | ctccatcccgggagcggtaccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F5 | tggcgcgccactagtggatccATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R5 | gtacatcccgggagcggtaccGAAAACTCCCCCAGACATTGAA |
表1 本研究所用引物
Table 1 Primers used in this study
| 用途 Application | 引物名称Primer name | 引物序列Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene cloning | RcERF4-F1 | ATGGCACCGAGAACCGAGA |
| RcERF4-R1 | TCAGGCGAGTTCCGCCGGA | |
| RcRAP2-12-F1 | ATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R1 | GAAAACTCCCCCAGACATTG | |
实时荧光定量PCR Quantitative real-time PCR | RcERF4-F2 | ACCACGGCTAAACCTAACGG |
| RcERF4-R2 | ATTCACATTCTCCGACGGCA | |
| RcRAP2-12-F2 | CGACTTGGTCTGGCCTGATT | |
| RcRAP2-12-R2 | TGCACTCCACGGATTTCACA | |
亚细胞定位和遗传转化烟草 Subcellular localization and genetic transformation of tobacco | RcERF4-F3 | aagtccggagctagctctagaATGGCACCGAGAACCGAGA |
| RcERF4-R3 | gcccttgctcaccatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F3 | aagtccggagctagctctagaATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R3 | gcccttgctcaccatggatccGAAAACTCCCCCAGACATTGAA | |
酵母双杂交 Yeast two-hybrid | RcERF4-F4 | gccatggaggccagtgaattcATGGCACCGAGAACCGAGA |
| RcERF4-R4 | cagctcgagctcgatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F4 | tcagaggaggacctgcatatgATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R4 | ctagttatgcggccgctgcagGAAAACTCCCCCAGACATTGAA | |
双分子荧光互补 Bimolecular fluorescence complementation | RcERF4-F5 | tggcgcgccactagtggatccATGGCACCGAGAACCGAGA |
| RcERF4-R5 | ctccatcccgggagcggtaccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F5 | tggcgcgccactagtggatccATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R5 | gtacatcccgggagcggtaccGAAAACTCCCCCAGACATTGAA |
| 名称 Name | 网址 Website | 用途 Application |
|---|---|---|
| ExPASy-ProtParam | https://web.expasy.org/protparam/ | 理化性质 Physicochemical properties |
| CDS | https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 结构域 Domain |
| TMHMM-2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | 跨膜结构 Transmembrane structure |
| SignalP-4.1 | https://services.healthtech.dtu.dk/services/SignalP-4.1/ | 信号肽 Signal peptide |
| NetGlycate-1.0 | https://services.healthtech.dtu.dk/services/NetGlycate-1.0/ | 糖基化位点 Glycosylation site |
| NetPhos-3.1 | https://services.healthtech.dtu.dk/services/NetPhos-3.1/ | 磷酸化位点 Phosphorylation site |
表2 生物信息学分析网址及用途
Table 2 Bioinformatics analysis websites and their applications
| 名称 Name | 网址 Website | 用途 Application |
|---|---|---|
| ExPASy-ProtParam | https://web.expasy.org/protparam/ | 理化性质 Physicochemical properties |
| CDS | https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 结构域 Domain |
| TMHMM-2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | 跨膜结构 Transmembrane structure |
| SignalP-4.1 | https://services.healthtech.dtu.dk/services/SignalP-4.1/ | 信号肽 Signal peptide |
| NetGlycate-1.0 | https://services.healthtech.dtu.dk/services/NetGlycate-1.0/ | 糖基化位点 Glycosylation site |
| NetPhos-3.1 | https://services.healthtech.dtu.dk/services/NetPhos-3.1/ | 磷酸化位点 Phosphorylation site |

图4 RcERF4(A)和RcRAP2-12(B)氨基酸序列比对Aa:蕨麻,XP_050371303.1;Ej:枇杷,AKN10304.1;Fa:草莓,AZL19463.1;Fv:野草莓,XP_004293846.1;Mp:苹果,NP_001385262.1;Ms:欧洲野苹果,XP_050108098.1;Pp:桃,ALO81027.1;Ra:尖齿黑莓,KAK9936415.1;Rc:月季,XP_024158160.1;Rr:玫瑰,XP_062013106.1;Pa:欧洲甜樱桃,XP_021819942.1; Pb:白梨,XP_009338318.1;Ps:山樱花,APY26916.1。红色圆点表示WLG元件;红色方块表示目标蛋白;红色框表示第14位和第19位氨基酸;蓝色框表示AP2结构域
Fig. 4 Amino acid sequence alignment of RcERF4 (A) and RcRAP2-12 (B)Aa: Argentina anserina, XP_050371303.1. Ej: Eriobotrya japonica, AKN10304.1. Fa: Fragaria × ananassa, AZL19463.1. Fv: Fragaria vesca subsp. vesca, XP_004293846.1. Mp: Malus pumila, NP_001385262.1. Ms: Malus sylvestris, XP_050108098.1. Pp: Prunus persica, ALO81027.1. Ra: Rubus argutus, KAK9936415.1. Rc: Rosa chinensis, XP_024158160.1. Rr: Rosa rugosa, XP_062013106.1. Pa: Prunus avium, XP_021819942.1. Pb: Pyrus × bretschneideri, XP_009338318.1. Ps: Prunus serrulata, APY26916.1. The red dot indicates the WLG component. The red square indicates the target protein. The red box indicates the 14th and 19th amino acids. The blue box indicates the AP2 domain
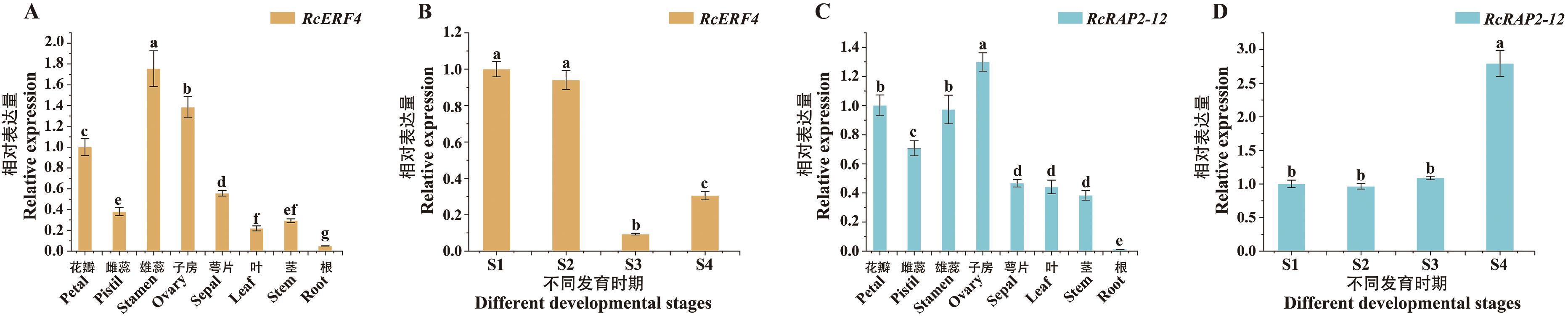
图6 RcERF4和RcRAP2-12在‘夏洛特夫人’不同组织部位和不同花发育时期的表达模式使用3次生物学重复;误差棒表示标准偏差;采用单因素方差进行统计分析,不同小写字母表示差异显著(P<0.05),下同
Fig. 6 Expression patterns of RcERF4 and RcRAP2-12 in different tissues and various floral developmental stages of 'Lady of Shalott'Three biological replicates were used. The error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA, different lower case letters indicate significant differences (P<0.05), the same below
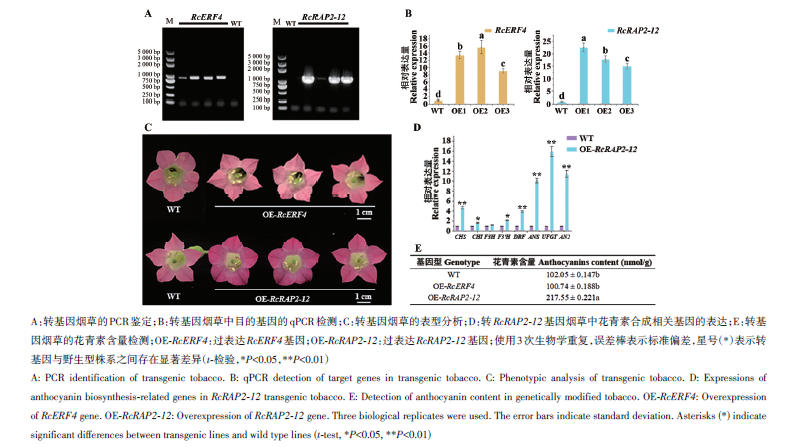
图8 转基因烟草的鉴定及分析A:转基因烟草的PCR鉴定;B:转基因烟草中目的基因的qPCR检测;C:转基因烟草的表型分析;D:转RcRAP2-12基因烟草中花青素合成相关基因的表达;E:转基因烟草的花青素含量检测;OE-RcERF4:过表达RcERF4基因;OE-RcRAP2-12:过表达RcRAP2-12基因;使用3次生物学重复,误差棒表示标准偏差,星号(*)表示转基因与野生型株系之间存在显著差异(t-检验,*P<0.05,**P<0.01)
Fig. 8 Identification and analysis of transgenic tobaccoA: PCR identification of transgenic tobacco. B: qPCR detection of target genes in transgenic tobacco. C: Phenotypic analysis of transgenic tobacco. D: Expressions of anthocyanin biosynthesis-related genes in RcRAP2-12 transgenic tobacco. E: Detection of anthocyanin content in genetically modified tobacco. OE-RcERF4: Overexpression of RcERF4 gene. OE-RcRAP2-12: Overexpression of RcRAP2-12 gene. Three biological replicates were used. The error bars indicate standard deviation. Asterisks (*) indicate significant differences between transgenic lines and wild type lines (t-test, *P<0.05, **P<0.01)
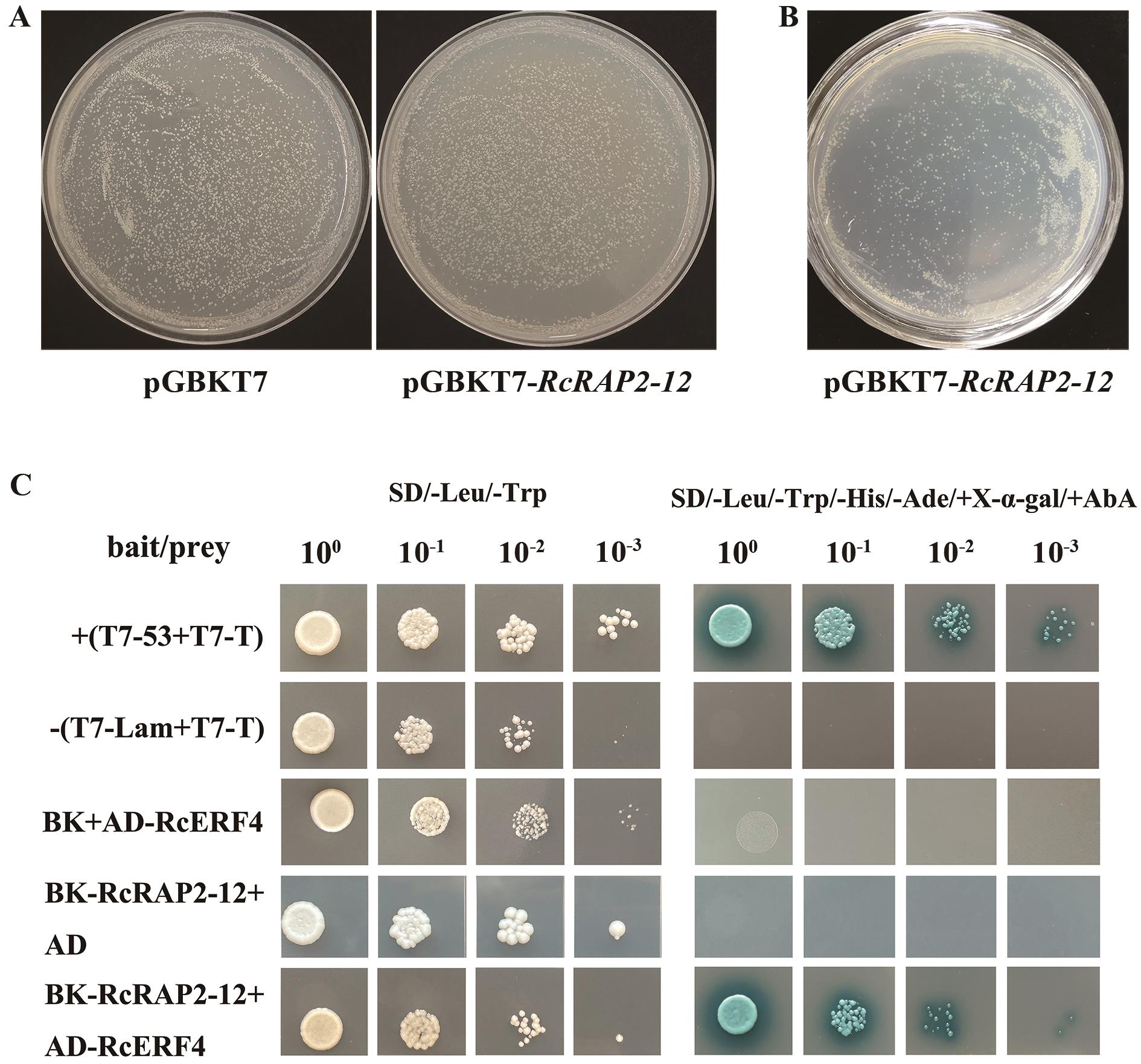
图9 酵母双杂交A:pGBKT7-RAP2-12的毒性检测;B:pGBKT7-RAP2-12的自激活验证;C:pGBKT7-RAP2-12和pGADT7-RcERF4的互作验证
Fig. 9 Yeast two-hybrid verificationA: Toxicity testing of pGBKT7-RAP2-12. B: Self-activation verification of pGBKT7-RAP2-12. C: Interaction validation between pGBKT7-RAP2-12 and pGADT7-RcERF4
| [1] | Ma ZM, Jin YM, Wu T, et al. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice [J]. Front Plant Sci, 2022, 13: 1007811. |
| [2] | Li HY, Du HP, Huang ZR, et al. The AP2/ERF transcription factor TOE4b regulates photoperiodic flowering and grain yield per plant in soybean [J]. Plant Biotechnol J, 2023, 21(8): 1682-1694. |
| [3] | Shi HY, Qanmber G, Yang ZR, et al. An AP2/ERF transcription factor GhERF109 negatively regulates plant growth and development in cotton [J]. Plant Sci, 2025, 352: 112365. |
| [4] | Zhang Y, Ji AJ, Xu ZC, et al. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza [J]. Plant Mol Biol, 2019, 100(1/2): 83-93. |
| [5] | Zhang J, Liu ZY, Sakamoto S, et al. ETHYLENE RESPONSE FACTOR 34 promotes secondary cell wall thickening and strength of rice peduncles [J]. Plant Physiol, 2022, 190(3): 1806-1820. |
| [6] | Xu L, Yang LJ, Li AP, et al. An AP2/ERF transcription factor confers chilling tolerance in rice [J]. Sci Adv, 2024, 10(35): eado4788. |
| [7] | Ritonga FN, Ngatia JN, Wang YR, et al. AP2/ERF, an important cold stress-related transcription factor family in plants: a review [J]. Physiol Mol Biol Plants, 2021, 27(9): 1953-1968. |
| [8] | Su ZL, Li AM, Wang M, et al. The role of AP2/ERF transcription factors in plant responses to biotic stress [J]. Int J Mol Sci, 2025, 26(10): 4921. |
| [9] | 兰孟焦, 后猛, 肖满秋, 等. AP2/ERF转录因子参与植物次生代谢和逆境胁迫响应的研究进展 [J]. 植物遗传资源学报, 2023, 24(5): 1223-1235. |
| Lan MJ, Kou M, Xiao MQ, et al. Research progress of AP2/ERF transcription factors participating in plant secondary metabolism and stress response [J]. J Plant Genet Resour, 2023, 24(5): 1223-1235. | |
| [10] | 卢勇杰, 夏海乾, 李永铃, 等. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析 [J]. 生物技术通报, 2025, 41(4): 266-277. |
| Lu YJ, Xia HQ, Li YL, et al. Cloning and expression analysis of AP2/ERF transcription factor NtESR2 in Nicotiana tabacum [J]. Biotechnol Bull, 2025, 41(4): 266-277. | |
| [11] | Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression [J]. Biochem Biophys Res Commun, 2002, 290(3): 998-1009. |
| [12] | Khaskheli AJ, Ahmed W, Ma C, et al. RhERF113 functions in ethylene-induced petal senescence by modulating cytokinin content in rose [J]. Plant Cell Physiol, 2018, 59(12): 2442-2451. |
| [13] | An JP, Zhang XW, Bi SQ, et al. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple [J]. Plant J, 2020, 101(3): 573-589. |
| [14] | Zhang SY, Liu S, Ren YS, et al. The ERF transcription factor ZbERF3 promotes ethylene-induced anthocyanin biosynthesis in Zanthoxylum bungeanum [J]. Plant Sci, 2024, 349: 112264. |
| [15] | Mo RL, Han GM, Zhu ZX, et al. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘Zijin’ mulberry fruits by interacting with MYBA and F3H genes [J]. Int J Mol Sci, 2022, 23(14): 7615. |
| [16] | Cao YW, Song M, Bi MM, et al. Lily (Lilium spp.) LhERF4 negatively affects anthocyanin biosynthesis by suppressing LhMYBSPLATTER transcription [J]. Plant Sci, 2024, 342: 112026. |
| [17] | Sun HY, Hu KD, Wei SW, et al. ETHYLENE RESPONSE FACTORS 4.1/4.2 with an EAR motif repress anthocyanin biosynthesis in red-skinned pears [J]. Plant Physiol, 2023, 192(3): 1892-1912. |
| [18] | Jiang SH, Wang HH, Zhang R, et al. Transcriptomic-based analysis to identify candidate genes for blue color rose breeding [J]. Plant Mol Biol, 2023, 111(4/5): 439-454. |
| [19] | Su MY, Damaris RN, Hu ZR, et al. Metabolomic analysis on the petal of ‘Chen Xi’ rose with light-induced color changes [J]. Plants, 2021, 10(10): 2065. |
| [20] | Ji NZ, Wang QY, Li SS, et al. Metabolic profile and transcriptome reveal the mystery of petal blotch formation in rose [J]. BMC Plant Biol, 2023, 23(1): 46. |
| [21] | 王超林. RcGT1和Rc3GGT在月季粉色系花瓣呈色过程中的功能研究 [D]. 北京: 北京农学院, 2023. |
| Wang CL. Function of RcGT1 and Rc3GGT for petal coloration in pink Rosa chinensis [D]. Beijing: Beijing University Of Agriculture, 2023. | |
| [22] | Yan HJ, Zhang H, Wang QG, et al. The Rosa chinensis cv. viridiflora phyllody phenotype is associated with misexpression of flower organ identity genes [J]. Front Plant Sci, 2016, 7: 996. |
| [23] | Gallois P, Marinho P. Leaf disk transformation using Agrobacterium tumefaciens -expression of heterologous genes in tobacco [J]. Methods Mol Biol, 1995, 49: 39-48. |
| [24] | 杨娟. 基于多组学分析的燕子花花色变异分子机理 [D]. 哈尔滨: 东北林业大学, 2023. |
| Yang J. The molecular mechanism of flower color variation of iris laevigata based on multi-omics analysis [D]. Harbin: Northeast Forestry University, 2023. | |
| [25] | Wang Z, Song GW, Zhang FJ, et al. Functional characterization of AP2/ERF transcription factors during flower development and anthocyanin biosynthesis related candidate genes in Lycoris [J]. Int J Mol Sci, 2023, 24(19): 14464. |
| [26] | Li DL, He YJ, Li SH, et al. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.) [J]. Plant Physiol Biochem, 2021, 167: 492-503. |
| [27] | Wang Z, Yang JC, Gao Q, et al. The transcription factor NtERF13a enhances abiotic stress tolerance and phenylpropanoid compounds biosynthesis in tobacco [J]. Plant Sci, 2023, 334: 111772. |
| [28] | He GR, Zhang R, Jiang SH, et al. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis [J]. Hortic Res, 2023, 10(6): uhad080. |
| [29] | Wang YG, Zhou LJ, Wang YX, et al. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of Chrysanthemum [J]. Sci Hortic, 2022, 293: 110674. |
| [30] | Zhou LJ, Wang YX, Wang YG, et al. Transcription factor CmbHLH16 regulates petal anthocyanin homeostasis under different lights in Chrysanthemum [J]. Plant Physiol, 2022, 190(2): 1134-1152. |
| [31] | Ji XL, Zhao LL, Liu BY, et al. MdZFP7 integrates JA and GA signals via interaction with MdJAZ2 and MdRGL3a in regulating anthocyanin biosynthesis and undergoes degradation by the E3 ubiquitin ligase MdBRG3 [J]. J Integr Plant Biol, 2025, 67(5): 1339-1363. |
| [1] | 曾厅, 张兰, 罗睿. 转录因子MpR2R3-MYB17调控地钱胞芽发育的功能研究[J]. 生物技术通报, 2026, 42(1): 208-217. |
| [2] | 吕呈聪, 衡蒙, 陈思琪, 金雪花. 彩色马蹄莲花青素苷转运相关ZhGSTF的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 161-169. |
| [3] | 任云儿, 伍国强, 成斌, 魏明. 甜菜BvATGs基因家族全基因组鉴定及盐胁迫下表达模式分析[J]. 生物技术通报, 2026, 42(1): 184-197. |
| [4] | 吴翠翠, 陈登科, 兰刚, 夏芝, 李朋波. 花生转录因子AhHDZ70的生物信息学分析及耐盐耐旱性研究[J]. 生物技术通报, 2026, 42(1): 198-207. |
| [5] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [6] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [7] | 张超超, 韩开元, 王彤, 陈仲. 毛白杨PtoYABBY2和PtoYABBY12的克隆及功能分析[J]. 生物技术通报, 2025, 41(9): 256-264. |
| [8] | 史发超, 姜永华, 刘海伦, 文英杰, 严倩. 荔枝LcTFL1基因的克隆与功能分析[J]. 生物技术通报, 2025, 41(9): 159-167. |
| [9] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [10] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [11] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [12] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [13] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [14] | 李玉珍, 李梦丹, 张蔚, 彭婷. 基于月季扩展蛋白基因家族鉴定的野蔷薇RmEXPB2基因功能研究[J]. 生物技术通报, 2025, 41(9): 182-194. |
| [15] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||