








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 182-194.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0216
• 研究报告 • 上一篇
收稿日期:2025-03-02
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
彭婷,女,博士,高级实验师,研究方向:观赏植物遗传育种与生物技术;E-mail: tpeng@mail.hzau.edu.cn作者简介:李玉珍,女,硕士研究生,研究方向 :观赏植物遗传育种与生物技术;E-mail: 1115662345@qq.com基金资助:
LI Yu-zhen1( ), LI Meng-dan1(
), LI Meng-dan1( ), ZHANG Wei1, PENG Ting2(
), ZHANG Wei1, PENG Ting2( )
)
Received:2025-03-02
Published:2025-09-26
Online:2025-09-24
摘要:
目的 系统鉴定中国古老月季‘月月粉’(Rosa chinensis ‘Old Blush’)扩展蛋白(Expansin)基因家族,解析其特征与表达模式,挖掘皮刺发育相关基因,并基于该结果进一步探究野蔷薇(Rosa multiflora)RmEXPB2基因的生物学功能,为深入鉴定扩展蛋白基因在蔷薇属植物皮刺形成中的功能奠定基础。 方法 利用生物信息学鉴定‘月月粉’基因组中的RcEXP,对其编码蛋白的氨基酸数量范围以及在染色体上的分布状况进行分析。通过构建系统进化树,剖析拟南芥(Arabidopsis thaliana)、‘月月粉’月季和森林草莓(Fragari avesca)的扩展蛋白(AtEXPs、RcEXPs和FvEXPs)的进化关系。检测‘月月粉’不同组织RcEXP表达模式及皮刺转录组中RcEXP表达。以皮刺研究模式植物野蔷薇作为实验材料,分离出候选基因RmEXPB2,研究该基因在皮刺发育进程中的表达变化情况,通过拟南芥异源表达探索其生物学功能。 结果 在‘月月粉’月季基因组中共鉴定出29个RcEXP,其编码蛋白含243‒313个氨基酸,在7条染色体上不均匀分布。系统进化树分析将AtEXPs、RcEXPs和FvEXPs分为4个亚组。在不同组织器官中,15个RcEXP表达模式各异,其中RcEXPB2和RcEXPA4在皮刺组织中优势表达。皮刺转录组数据中筛选获得13个RcEXP,在皮刺发育的特定阶段显著表达。从野蔷薇中分离出候选基因RmEXPB2,其在皮刺中优势表达,并且在皮刺发育末期表达量最高。异源表达RmEXPB2显著促进转基因拟南芥叶片扩展与根毛生长。 结论 月季‘月月粉’基因组中共鉴定出29个RcEXPs,其中13个成员在皮刺发育的特定阶段显著表达。野蔷薇RmEXPB2是一个皮刺优势表达基因,可能在皮刺伸长过程中发挥积极作用。
李玉珍, 李梦丹, 张蔚, 彭婷. 基于月季扩展蛋白基因家族鉴定的野蔷薇RmEXPB2基因功能研究[J]. 生物技术通报, 2025, 41(9): 182-194.
LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp.[J]. Biotechnology Bulletin, 2025, 41(9): 182-194.
| Gene name | Forward primer (5′‒3′) | Reverse primer (5′‒3′) |
|---|---|---|
| RmUBC | GCCAGAGATTGCCCATATGTA | TCACAGAGTCCTAGCAGCACA |
| AtEF1α | GCAAGATGGATGCCACTACCC | GCAAGATGGATGCCACTACCC |
| RmEXPB2 | TCTAGAATGGCTAGTTTAAGAAGCAT | CCCGGGTAGATAATTGACCACGGATC |
| RmEXPB2-qRT | TGCTGCTTGTTTGAGCAGAG | AAGCTCCACATTCCCTGCC |
| RmEXPB2-eGFP | GAGAACACGGGGGACTCTAGAATGGCTAGTTTAAGAAGCAT | GCCCTTGCTCACCATCCCGGGTAGATAATTGACCACGGATC |
表1 RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR
| Gene name | Forward primer (5′‒3′) | Reverse primer (5′‒3′) |
|---|---|---|
| RmUBC | GCCAGAGATTGCCCATATGTA | TCACAGAGTCCTAGCAGCACA |
| AtEF1α | GCAAGATGGATGCCACTACCC | GCAAGATGGATGCCACTACCC |
| RmEXPB2 | TCTAGAATGGCTAGTTTAAGAAGCAT | CCCGGGTAGATAATTGACCACGGATC |
| RmEXPB2-qRT | TGCTGCTTGTTTGAGCAGAG | AAGCTCCACATTCCCTGCC |
| RmEXPB2-eGFP | GAGAACACGGGGGACTCTAGAATGGCTAGTTTAAGAAGCAT | GCCCTTGCTCACCATCCCGGGTAGATAATTGACCACGGATC |
基因ID Gene ID | 基因 Gene | 染色体位置 Chr location | 等电点 pI | 分子量 Molecular weight (Da) | 蛋白长度 Protein length (aa) | 亚细胞定位Subcellular localization | 信号肽长度Signal peptie length (aa) |
|---|---|---|---|---|---|---|---|
| RchiOBHmChr5g0072761 | RcEXPA1 | Chr5 | 8.12 | 27 145.26 | 254 | Cell wall | 26 |
| RchiOBHmChr7g0178811 | RcEXPA2 | Chr7 | 8.82 | 28 302.47 | 256 | Cell wall | 22 |
| RchiOBHmChr7g0199891 | RcEXPA3 | Chr7 | 9.55 | 29 064.3 | 262 | Chloroplast | 28 |
| RchiOBHmChr7g0181621 | RcEXPA4 | Chr7 | 8.64 | 26 524.79 | 250 | Cell wall | 20 |
| RchiOBHmChr6g0281211 | RcEXPA5 | Chr6 | 6.93 | 27 816.75 | 253 | Cell wall | 23 |
| RchiOBHmChr3g0488771 | RcEXPA6 | Chr3 | 7.55 | 29 302.32 | 272 | Cell wall | 24 |
| RchiOBHmChr3g0470931 | RcEXPA7 | Chr3 | 9.00 | 28 950.9 | 260 | Chloroplast | 24 |
| RchiOBHmChr4g0439541 | RcEXPA8 | Chr4 | 9.56 | 31 940.33 | 297 | Mitochondrion | |
| RchiOBHmChr3g0470941 | RcEXPA9 | Chr3 | 5.68 | 29 434.08 | 266 | Cell wall | 24 |
| RchiOBHmChr5g0033911 | RcEXPA10 | Chr5 | 9.69 | 27 795.93 | 255 | Cell wall | 30 |
| RchiOBHmChr1g0372131 | RcEXPA11 | Chr1 | 7.99 | 26 919.1 | 254 | Cell wall | 25 |
| RchiOBHmChr3g0470961 | RcEXPA12 | Chr3 | 9.68 | 31 679.04 | 286 | Chloroplast | 25 |
| RchiOBHmChr6g0245581 | RcEXPA13 | Chr6 | 5.22 | 28 332.98 | 260 | Cell wall | 27 |
| RchiOBHmChr1g0367791 | RcEXPA14 | Chr1 | 9.53 | 27 917.86 | 259 | Chloroplast | 22 |
| RchiOBHmChr3g0466461 | RcEXPA15 | Chr3 | 9.50 | 27 787.56 | 259 | Cell wall | 21 |
| RchiOBHmChr2g0166091 | RcEXPA16 | Chr2 | 8.10 | 27 554.38 | 250 | Chloroplast | 22 |
| RchiOBHmChr5g0065301 | RcEXPA17 | Chr5 | 9.68 | 27 890.68 | 260 | Cell wall | 22 |
| RchiOBHmChr3g0460691 | RcEXPA18 | Chr3 | 8.36 | 27 487.08 | 255 | Cell wall | 24 |
| RchiOBHmChr6g0303551 | RcEXPA19 | Chr6 | 8.87 | 27 306.82 | 257 | Chloroplast | 23 |
| RchiOBHmChr2g0140121 | RcEXPA20 | Chr2 | 8.39 | 35 186.7 | 313 | Cell wall | 21 |
| RchiOBHmChr2g0167701 | RcEXPA21 | Chr2 | 9.35 | 26 293.59 | 243 | Cell wall | 20 |
| RchiOBHmChr2g0166111 | RcEXPA22 | Chr2 | 6.06 | 27 581.6 | 253 | Cell wall | 23 |
| RchiOBHmChr5g0084081 | RcEXPA23 | Chr5 | 8.85 | 27 781.15 | 261 | Nucleus | 22 |
| RchiOBHmChr2g0126231 | RcEXPB1 | Chr2 | 8.73 | 27 394.3 | 255 | Cell wall | 23 |
| RchiOBHmChr1g0373921 | RcEXPB2 | Chr1 | 4.97 | 28 762.08 | 273 | Chloroplast | 23 |
| RchiOBHmChr1g0348851 | RcEXPB3 | Chr1 | 6.40 | 30 068.99 | 287 | Chloroplast | 29 |
| RchiOBHmChr6g0279781 | RcEXLA1 | Chr6 | 8.8 | 30 176.7 | 278 | Chloroplast | |
| RchiOBHmChr7g0205401 | RcEXLB1 | Chr7 | 6.87 | 27 333.67 | 250 | Cell wall | 22 |
| RchiOBHmChr7g0208241 | RcEXLB2 | Chr7 | 4.70 | 28 099.57 | 258 | Chloroplast | 25 |
表2 ‘月月粉’月季扩展蛋白基因家族成员基本信息
Table 2 Basic information of expansin gene family members in R. chinensis'Old Blush'
基因ID Gene ID | 基因 Gene | 染色体位置 Chr location | 等电点 pI | 分子量 Molecular weight (Da) | 蛋白长度 Protein length (aa) | 亚细胞定位Subcellular localization | 信号肽长度Signal peptie length (aa) |
|---|---|---|---|---|---|---|---|
| RchiOBHmChr5g0072761 | RcEXPA1 | Chr5 | 8.12 | 27 145.26 | 254 | Cell wall | 26 |
| RchiOBHmChr7g0178811 | RcEXPA2 | Chr7 | 8.82 | 28 302.47 | 256 | Cell wall | 22 |
| RchiOBHmChr7g0199891 | RcEXPA3 | Chr7 | 9.55 | 29 064.3 | 262 | Chloroplast | 28 |
| RchiOBHmChr7g0181621 | RcEXPA4 | Chr7 | 8.64 | 26 524.79 | 250 | Cell wall | 20 |
| RchiOBHmChr6g0281211 | RcEXPA5 | Chr6 | 6.93 | 27 816.75 | 253 | Cell wall | 23 |
| RchiOBHmChr3g0488771 | RcEXPA6 | Chr3 | 7.55 | 29 302.32 | 272 | Cell wall | 24 |
| RchiOBHmChr3g0470931 | RcEXPA7 | Chr3 | 9.00 | 28 950.9 | 260 | Chloroplast | 24 |
| RchiOBHmChr4g0439541 | RcEXPA8 | Chr4 | 9.56 | 31 940.33 | 297 | Mitochondrion | |
| RchiOBHmChr3g0470941 | RcEXPA9 | Chr3 | 5.68 | 29 434.08 | 266 | Cell wall | 24 |
| RchiOBHmChr5g0033911 | RcEXPA10 | Chr5 | 9.69 | 27 795.93 | 255 | Cell wall | 30 |
| RchiOBHmChr1g0372131 | RcEXPA11 | Chr1 | 7.99 | 26 919.1 | 254 | Cell wall | 25 |
| RchiOBHmChr3g0470961 | RcEXPA12 | Chr3 | 9.68 | 31 679.04 | 286 | Chloroplast | 25 |
| RchiOBHmChr6g0245581 | RcEXPA13 | Chr6 | 5.22 | 28 332.98 | 260 | Cell wall | 27 |
| RchiOBHmChr1g0367791 | RcEXPA14 | Chr1 | 9.53 | 27 917.86 | 259 | Chloroplast | 22 |
| RchiOBHmChr3g0466461 | RcEXPA15 | Chr3 | 9.50 | 27 787.56 | 259 | Cell wall | 21 |
| RchiOBHmChr2g0166091 | RcEXPA16 | Chr2 | 8.10 | 27 554.38 | 250 | Chloroplast | 22 |
| RchiOBHmChr5g0065301 | RcEXPA17 | Chr5 | 9.68 | 27 890.68 | 260 | Cell wall | 22 |
| RchiOBHmChr3g0460691 | RcEXPA18 | Chr3 | 8.36 | 27 487.08 | 255 | Cell wall | 24 |
| RchiOBHmChr6g0303551 | RcEXPA19 | Chr6 | 8.87 | 27 306.82 | 257 | Chloroplast | 23 |
| RchiOBHmChr2g0140121 | RcEXPA20 | Chr2 | 8.39 | 35 186.7 | 313 | Cell wall | 21 |
| RchiOBHmChr2g0167701 | RcEXPA21 | Chr2 | 9.35 | 26 293.59 | 243 | Cell wall | 20 |
| RchiOBHmChr2g0166111 | RcEXPA22 | Chr2 | 6.06 | 27 581.6 | 253 | Cell wall | 23 |
| RchiOBHmChr5g0084081 | RcEXPA23 | Chr5 | 8.85 | 27 781.15 | 261 | Nucleus | 22 |
| RchiOBHmChr2g0126231 | RcEXPB1 | Chr2 | 8.73 | 27 394.3 | 255 | Cell wall | 23 |
| RchiOBHmChr1g0373921 | RcEXPB2 | Chr1 | 4.97 | 28 762.08 | 273 | Chloroplast | 23 |
| RchiOBHmChr1g0348851 | RcEXPB3 | Chr1 | 6.40 | 30 068.99 | 287 | Chloroplast | 29 |
| RchiOBHmChr6g0279781 | RcEXLA1 | Chr6 | 8.8 | 30 176.7 | 278 | Chloroplast | |
| RchiOBHmChr7g0205401 | RcEXLB1 | Chr7 | 6.87 | 27 333.67 | 250 | Cell wall | 22 |
| RchiOBHmChr7g0208241 | RcEXLB2 | Chr7 | 4.70 | 28 099.57 | 258 | Chloroplast | 25 |
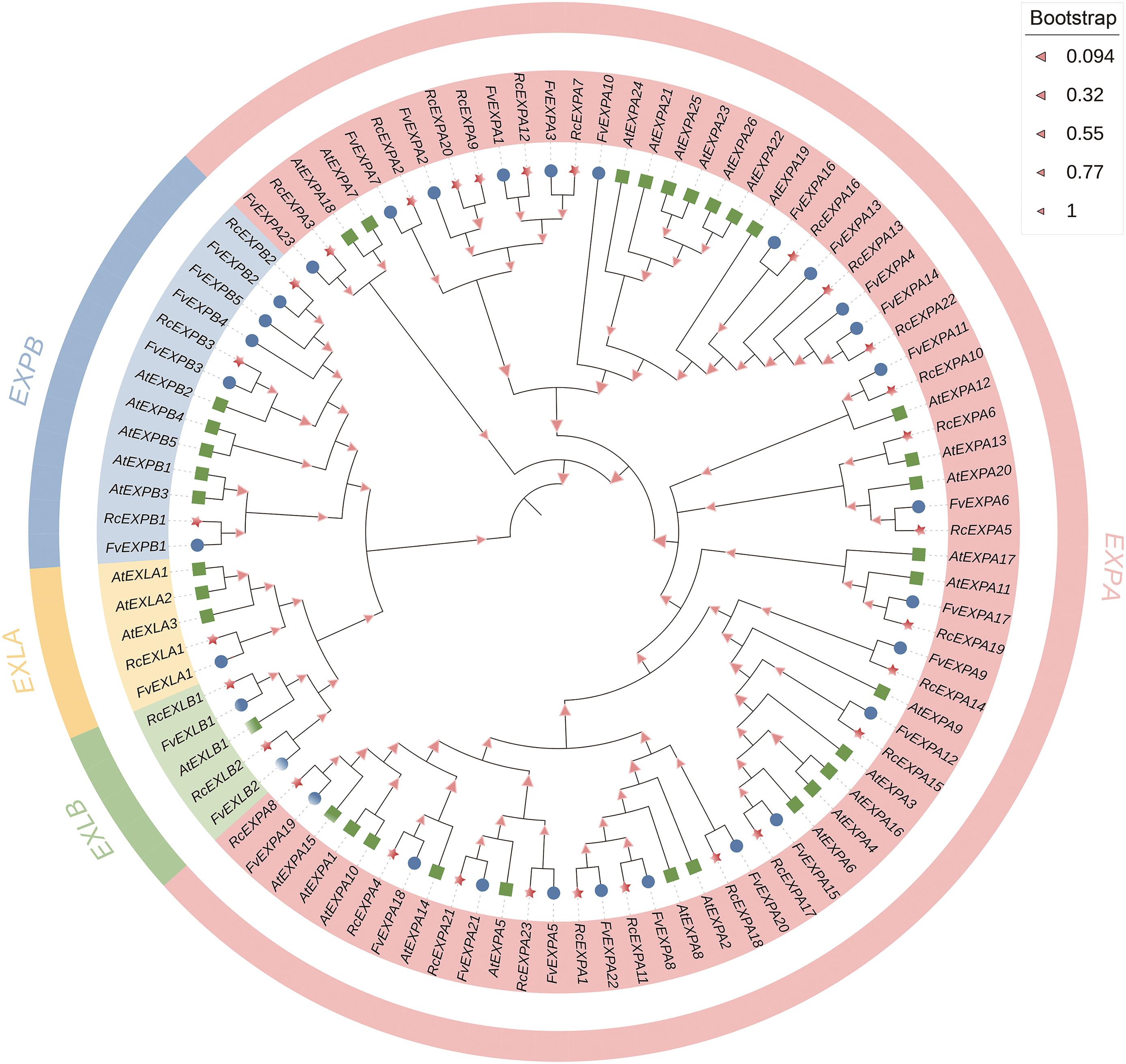
图2 ‘月月粉’月季、森林草莓和拟南芥的expansin基因家族进化树红色五角星代表‘月月粉’月季,蓝色圆形代表森林草莓,绿色方框代表拟南芥
Fig. 2 Phylogenetic tree of expansin gene family in R. chinensis'Old Blush', F. avesca and ArabidopsisThe red five-pointed star indicates R.chinensis 'Old Blush', the blue circle indicates F. avesca, and the green box indicates Arabidopsis

图3 ‘月月粉’月季expansin基因家族保守基序及基因结构分析A:保守基序分析;B:保守结构域分析;C:基因结构分析
Fig. 3 Analysis of conserved motif and gene structure of expansin gene family of R. chinensis'Old Blush'A: Conservative motif analysis. B: Conserved domain analysis. C: Gene structure analysis
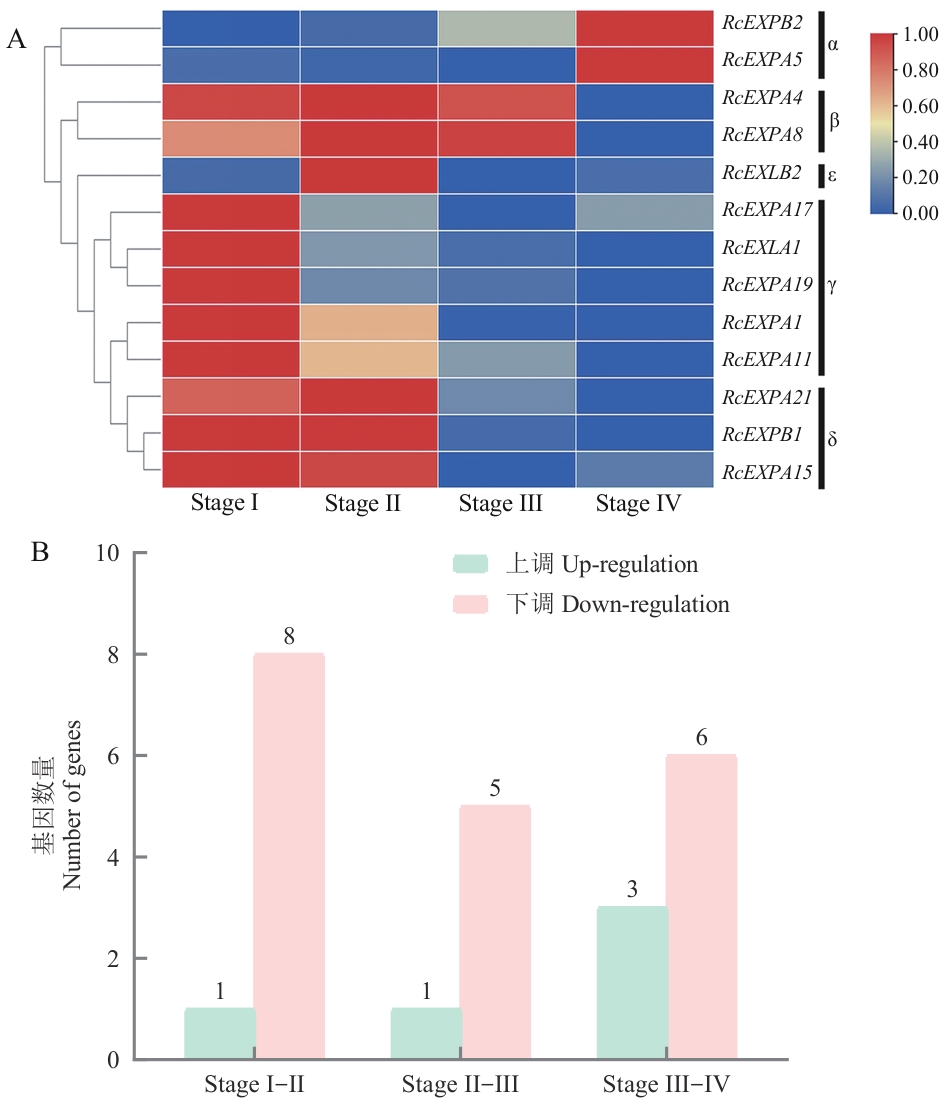
图6 ‘月月粉’月季部分expansin基因在不同发育阶段皮刺中的表达分析(A)及基因数量(B)Stage I:柔软幼嫩、尚未木质化的皮刺;Stage Ⅱ:一定程度木质化、尖端开始向基部弯曲的皮刺;Stage Ⅲ:木质化较完全、形态基本成熟的皮刺;Stage IV:发育成熟、并初步开始衰老的皮刺
Fig. 6 Expression pattern analysis (A) and the gene number (B) of some expansin genes in different developmental stages of prickles in R. chinensis'Old Blush'Stage I: Soft and tender prickles that have not yet lignified. Stage Ⅱ: Prickles that are lignified to a certain extent and whose tips begin to bend toward the base. Stage Ⅲ: Prickles with relatively complete lignification and basically mature morphology. Stage IV: Prickles that have matured and initially begun to age

图7 RmEXPB2的表达量分析A:RmEXPB2在野蔷薇不同组织中的表达水平;B:RmEXPB2在野蔷薇不同发育阶段皮刺组织中的表达水平;C:野生型及T1代转基因株系RmEXPB2表达量检测;D:野生型及T2代转基因株系RmEXPB2表达量检测;Stage I:野蔷薇起始期皮刺;Stage Ⅱ:快速生长期皮刺;Stage Ⅲ:过渡期皮刺;Stage IV:硬化期皮刺;Stage V成熟期皮刺;line 1‒10代表转基因株系1‒10,WT代表野生型。ns表示P>0.05;*P<0.05;**P<0.01;***P<0.001。下同
Fig. 7 Expression analysis of RmEXPB2A: Expressions of RmEXPB2 in different tissues of R. multiflora. B: Expressions of RmEXPB2 in the prickles at different developmental stages. C: RmEXPB2 expression levels in WT and RmEXPB2-OE (T1) Arabidopsis. D: RmEXPB2 expression levels in WT and RmEXPB2-OE (T2) Arabidopsis. Stage I: Starting period of R. multiflora prickles; Stage II: rapid growth period prickles; Stage III: transition period prickles; Stage IV: hardening period prickles; Stage V: maturation period prickles; line 1-10 indicates transgenic line 1‒10, WT indicates wild type. ns indicates P>0.05; * indicates P<0.05; ** indicates P<0.01; *** indicates P<0.001. The same below
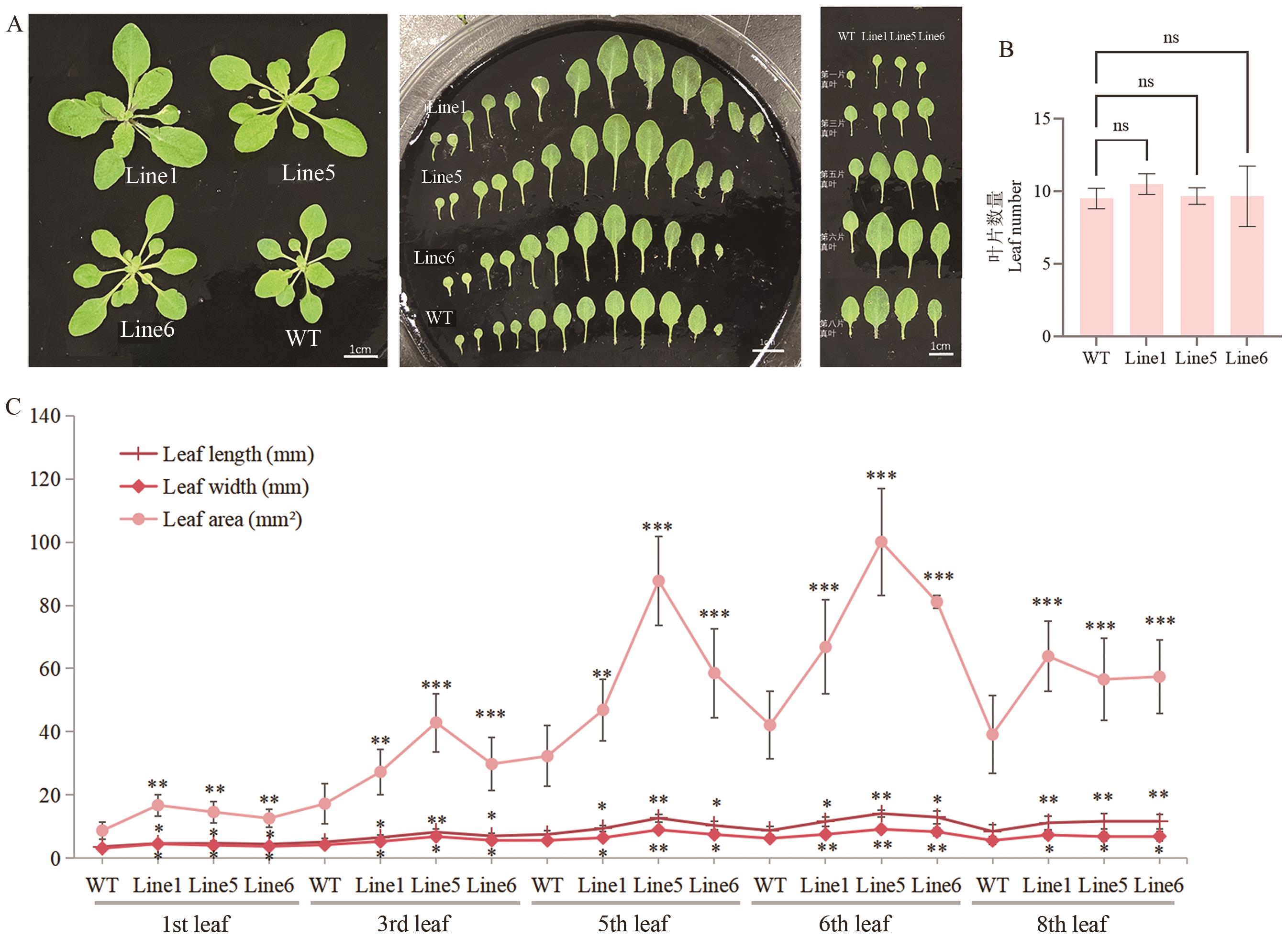
图8 RmEXPB2转基因拟南芥T2代株系的叶片表型A:莲座叶表型图;B:莲座叶数量,n>20;C:第1、3、5、6、8片莲座叶的叶长、叶宽、叶面积;Line 1、5、6代表转基因株系1、5、6,WT代表野生型。下同
Fig. 8 Leaf phenotypes of RmEXPB2-OE (T2) Arabidopsis plantsA: True leaf phenotypic diagram. B: Number of true leaves. n>20. C: The 1st, 3rd, 5th, 6th, 8th leaf length, width and area of the true leaf. Line1, 5, 6 indicates transgenic line1, 5, 6, WT indicates wild type. The same below

图9 RmEXPB2转基因拟南芥T2代植株的根部表型A:野生型及RmEXPB2转基因拟南芥的根;B:野生型及RmEXPB2转基因拟南芥的根毛;C‒D:野生型与RmEXPB2转基因拟南芥的主根长度、下胚轴长度;E‒F:野生型及RmEXPB2转基因拟南芥的根毛长度、根毛密度;n>20
Fig. 9 Root phenotypic analysis of RmEXPB2-OE (T2) Arabidopsis plantsA: Roots of WT and RmEXPB2-OE (T2) transgenic Arabidopsis. B: Root hairs of WT and RmEXPB2-OE (T2) Arabidopsis. C‒D: The main root length and hypocotyl length of WT and RmEXPB2-OE (T2) transgenic Arabidopsis. E‒F: The root hair length and root hair density of RmEXPB2-OE (T2) Arabidopsis. n>20
| [1] | Nakamura N, Hirakawa H, Sato S, et al. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses [J]. DNA Res, 2018, 25(2): 113-121. |
| [2] | Zhang Y, Zhao MJ, Zhu W, et al. Nonglandular prickle formation is associated with development and secondary metabolism-related genes in Rosa multiflora [J]. Physiol Plant, 2021, 173(3): 1147-1162. |
| [3] | McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants [J]. Plant Cell, 1992, 4(11): 1425-1433. |
| [4] | Arslan B, İncili ÇY, Ulu F, et al. Comparative genomic analysis of expansin superfamily gene members in zucchini and cucumber and their expression profiles under different abiotic stresses [J]. Physiol Mol Biol Plants, 2021, 27(12): 2739-2756. |
| [5] | Cosgrove DJ. Plant cell wall loosening by expansins [J]. Annu Rev Cell Dev Biol, 2024, 40(1): 329-352. |
| [6] | Wang ZZ, Cao JB, Lin N, et al. Origin, evolution, and diversification of the expansin family in plants [J]. Int J Mol Sci, 2024, 25(21): 11814. |
| [7] | Kök BÖ, Celik Altunoglu Y, Öncül AB, et al. Expansin gene family database: a comprehensive bioinformatics resource for plant expansin multigene family [J]. J Bioinform Comput Biol, 2023, 21(3): 2350015. |
| [8] | Liu WM, Lyu TQ, Xu LA, et al. Complex molecular evolution and expression of expansin gene families in three basic diploid species of Brassica [J]. Int J Mol Sci, 2020, 21(10): 3424. |
| [9] | Goh HH, Sloan J, Dorca-Fornell C, et al. Inducible repression of multiple expansin genes leads to growth suppression during leaf development [J]. Plant Physiol, 2012, 159(4): 1759-1770. |
| [10] | Yan A, Wu MJ, Yan LM, et al. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis [J]. PLoS One, 2014, 9(1): e85208. |
| [11] | Feng X, Li CT, He FM, et al. Genome-wide identification of expansin genes in wild soybean (Glycine soja) and functional characterization of Expansin B1 (GsEXPB1) in soybean hair root [J]. Int J Mol Sci, 2022, 23(10): 5407. |
| [12] | Liu WM, Xu LA, Lin H, et al. Two expansin genes, AtEXPA4 and AtEXPB5, are redundantly required for pollen tube growth and AtEXPA4 is involved in primary root elongation in Arabidopsis thaliana [J]. Genes, 2021, 12(2): 249. |
| [13] | Jiang FL, Lopez A, Jeon S, et al. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking [J]. Hortic Res, 2019, 6: 17. |
| [14] | Calderini DF, Castillo FM, Arenas-M A, et al. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential [J]. New Phytol, 2021, 230(2): 629-640. |
| [15] | Dai FW, Zhang CQ, Jiang XQ, et al. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals [J]. Plant Physiol, 2012, 160(4): 2064-2082. |
| [16] | Lü PT, Kang M, Jiang XQ, et al. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis [J]. Planta, 2013, 237(6): 1547-1559. |
| [17] | Raymond O, Gouzy J, Just J, et al. The Rosagenome provides new insights into the domestication of modern roses [J]. Nat Genet, 2018, 50(6): 772-777. |
| [18] | Han Y, Yu JY, Zhao T, et al. Dissecting the genome-wide evolution and function of R2R3-MYB transcription factor family in Rosa chinensis [J]. Genes, 2019, 10(10): 823. |
| [19] | 由玉婉, 张雨, 孙嘉毅, 等. ‘月月粉’月季NAC家族全基因组鉴定及皮刺发育相关成员的筛选 [J]. 中国农业科学, 2022, 55(24): 4895-4911. |
| You YW, Zhang Y, Sun JY, et al. Genome-wide identification of NAC family and screening of its members related to prickle development in Rosa chinensis old blush [J]. Sci Agric Sin, 2022, 55(24): 4895-4911. | |
| [20] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [21] | Hepler NK, Bowman A, Carey RE, et al. Expansin gene loss is a common occurrence during adaptation to an aquatic environment [J]. Plant J, 2020, 101(3): 666-680. |
| [22] | Krishnamurthy P, Hong JK, Kim JA, et al. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication [J]. Mol Genet Genomics, 2015, 290(2): 521-530. |
| [23] | Liu XP, Dong SY, Miao H, et al. Genome-wide analysis of expansins and their role in fruit spine development in cucumber (Cucumis sativus L.) [J]. Hortic Plant J, 2022, 8(6): 757-768. |
| [24] | Sampedro J, Carey RE, Cosgrove DJ. Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs [J]. J Plant Res, 2006, 119(1): 11-21. |
| [25] | Zhang W, Yan HW, Chen WJ, et al. Genome-wide identification and characterization of maize expansin genes expressed in endosperm [J]. Mol Genet Genomics, 2014, 289(6): 1061-1074. |
| [26] | Qiu DY, Xu SL, Wang Y, et al. Primary cell wall modifying proteins regulate wall mechanics to steer plant morphogenesis [J]. Front Plant Sci, 2021, 12: 751372. |
| [27] | Ono K. Signal peptides and their fragments in post-translation: novel insights of signal peptides [J]. Int J Mol Sci, 2024, 25(24): 13534. |
| [28] | Zhang JL, Dong TT, Zhu MK, et al. Transcriptome- and genome-wide systematic identification of expansin gene family and their expression in tuberous root development and stress responses in sweetpotato (Ipomoea batatas) [J]. Front Plant Sci, 2024, 15: 1412540. |
| [29] | Lou Y, Zhou HS, Han Y, et al. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana [J]. New Phytol, 2018, 217(1): 378-391. |
| [30] | Zhou NN, Simonneau F, Thouroude T, et al. Morphological studies of rose prickles provide new insights [J]. Hortic Res, 2021, 8(1): 221. |
| [31] | Swarnkar MK, Kumar P, Dogra V, et al. Prickle morphogenesis in rose is coupled with secondary metabolite accumulation and governed by canonical MBW transcriptional complex [J]. Plant Direct, 2021, 5(6): e00325. |
| [32] | Saint-Oyant LH, Ruttink T, Hamama L, et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits [J]. Nat Plants, 2018, 4(7): 473-484. |
| [33] | Xing SC, Li F, Guo QF, et al. The involvement of an expansin gene TaEXPB23 from wheat in regulating plant cell growth [J]. Biol Plant, 2009, 53(3): 429-434. |
| [34] | Zou HY, Wenwen YH, Zang GC, et al. OsEXPB2, a β-expansin gene, is involved in rice root system architecture [J]. Mol Breed, 2015, 35(1): 41. |
| [35] | Lin CF, Choi HS, Cho HT. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis [J]. Mol Cells, 2011, 31(4): 393-398. |
| [36] | Chen SK, Luo YX, Wang GJ, et al. Genome-wide identification of expansin genes in Brachypodium distachyon and functional characterization of BdEXPA27 [J]. Plant Sci, 2020, 296: 110490. |
| [1] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [2] | 黄旭升, 周雅莉, 柴旭东, 闻婧, 王计平, 贾小云, 李润植. 紫苏质体型PfLPAT1B基因的克隆及其在油脂合成中的功能分析[J]. 生物技术通报, 2025, 41(7): 226-236. |
| [3] | 王芳, 乔帅, 宋伟, 崔鹏娟, 廖安忠, 谭文芳, 杨松涛. 甘薯IbNRT2基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(7): 193-204. |
| [4] | 李彩霞, 李艺, 穆宏秀, 林俊轩, 白龙强, 孙美华, 苗妍秀. 中国南瓜bHLH转录因子家族的鉴定与生物信息学分析[J]. 生物技术通报, 2025, 41(1): 186-197. |
| [5] | 林彤, 袁程, 董陈文华, 曾孟琼, 杨燕, 毛自朝, 林春. 藜麦配子发育相关基因CqSTK的筛选及功能分析[J]. 生物技术通报, 2024, 40(8): 83-94. |
| [6] | 闫欢欢, 尚怡彤, 王丽红, 田学琴, 廖海艳, 曾斌, 胡志宏. 米曲霉异源表达合成虫草素[J]. 生物技术通报, 2024, 40(6): 290-298. |
| [7] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [8] | 殷亮, 王代玮, 刘悦莹, 刘海燕, 罗光宏. 蛋白酶SpP1基因克隆、表达及酶学性质的表征[J]. 生物技术通报, 2024, 40(4): 278-286. |
| [9] | 郑菲, 杨俊钊, 牛羽丰, 李蕊麟, 赵国柱. 嗜热毁丝菌裂解性多糖单加氧酶TtLPMO9I的酶学性质及其功能研究[J]. 生物技术通报, 2024, 40(2): 289-299. |
| [10] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [11] | 马玉倩, 孙东辉, 岳浩峰, 辛佳瑜, 刘宁, 曹志艳. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135. |
| [12] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [13] | 牛馨, 张莹, 王茂军, 刘文龙, 路福平, 李玉. 解淀粉芽胞杆菌不同整合位点对外源碱性蛋白酶表达的影响[J]. 生物技术通报, 2022, 38(4): 253-260. |
| [14] | 王玥, 高庆华, 董聪, 罗同阳, 王庆庆. 密码子优化的吡喃糖氧化酶基因在毕赤酵母中的表达[J]. 生物技术通报, 2022, 38(4): 269-277. |
| [15] | 王博雅, 姜勇, 黄艳, 曹颖, 胡尚连. 慈竹纤维素合酶BeCesA4的克隆及功能分析[J]. 生物技术通报, 2022, 38(11): 185-193. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||