








生物技术通报 ›› 2021, Vol. 37 ›› Issue (5): 56-66.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0919
收稿日期:2020-07-22
出版日期:2021-05-26
发布日期:2021-06-11
作者简介:邱小宇,男,硕士研究生,研究方向:动物肉品质调控;E-mail: 基金资助:
QIU Xiao-yu1( ), LIU Zuo-hua1,2, QI Ren-li1,3(
), LIU Zuo-hua1,2, QI Ren-li1,3( )
)
Received:2020-07-22
Published:2021-05-26
Online:2021-06-11
摘要:
通过比较无菌仔猪和普通仔猪在生长早期脂肪沉积的差异及脂肪组织基因转录表达谱的不同,评估肠道微生物定植对猪脂肪组织早期发育的直接影响。分别采集25日龄无菌仔猪(3头)和普通仔猪(3头)的颈部皮下脂肪,切片后经HE染色观察不同猪脂肪细胞的形态差异;Western Blot检测脂肪合成调控因子的蛋白表达差异;ELISA法测定不同猪脂肪组织分泌产生的脂肪细胞因子含量;转录组测序(RNA-seq)分析不同猪脂肪组织中的基因表达谱,鉴定关键差异基因及其相关信号网络。与普通仔猪相比,相同日龄的无菌仔猪体脂沉积量较少,皮下脂肪厚度和脂肪细胞尺寸均明显减小(P<0.001);脂肪组织中的脂肪酸结合蛋白4(fatty acid binding protein 4,FABP4)、过氧化物酶体增殖体激活受体γ(peroxisome proliferators activated recepor γ,PPARγ)、乙酰辅酶A羧化酶(acetyl CoA carboxylase,ACC)、脂肪酸合成酶(fatty acid synthase,FAS)的蛋白表达水平显著降低(P<0.05)。脂肪产生的功能性细胞因子脂联素(adiponectin,P=0.100)和瘦素(leptin,P=0.095)有着不同程度的减少。测序结果显示与普通猪相比,无菌猪的脂肪中有695个基因显著差异表达(P<0.05),包括338个基因表达上调,357个基因表达下调。qRT-PCR验证了转录组测序结果的准确性。KEGG通路富集分析结果显示差异基因主要与脂质合成代谢、分解代谢、脂质氧化、能量稳态相关。肠道微生物的定植显著影响了动物脂肪组织的发育、代谢和生理功能。
邱小宇, 刘作华, 齐仁立. 无菌猪和普通猪早期脂肪发育及脂肪组织基因转录表达的差异[J]. 生物技术通报, 2021, 37(5): 56-66.
QIU Xiao-yu, LIU Zuo-hua, QI Ren-li. Differences in Early Fat Development and Gene Transcription Expression in the Adipose Tissues of Piglets with and Without Gut Microbiota[J]. Biotechnology Bulletin, 2021, 37(5): 56-66.

图1 无菌仔猪和普通仔猪脂肪沉积的差异 A:无菌仔猪和普通仔猪脂肪组织HE染色;B:无菌仔猪和普通仔猪颈部皮下脂肪厚度;C:无菌仔猪和普通仔猪脂肪组织中脂肪细胞直径。*** P<0.001;**** P<0.0001
Fig. 1 Difference of fat deposition between GF pigs and normal pigs A: He staining of fat tissue of GF pigs and normal pigs. B: Back-fat of GF pigs and Normal pigs. C: Diameter of fat cells in fat tissue of GF pigs and Normal pigs. *** P<0.001;**** P<0.0001

图2 无菌猪和普通猪的脂肪合成调控分子表达和脂肪细胞因子含量的差异 A:无菌仔猪和普通仔猪脂肪组织FABP4、PPARγ、ACC、FAS的差异表达;B:无菌仔猪和普通仔猪脂肪组中的脂联素(Adiponection)的含量;C:无菌仔猪和普通仔猪脂肪组织瘦素(Leptin)的含量。*P<0.05;**P<0.01
Fig.2 Differences in fat synthesis regulatory factors and adipocytokines between GF pigs and normal pigs A: The differential expression of FABP4, PPAR γ, ACC and FAS in adipose tissue of GF and Normal piglets. B: The differential secretion of adiponectin. C: The differential secretion of adiponectin leptin. * P<0.05;** P<0.01
| 样品名 Sample name | 总序列数 Raw reads | 干净序列数 Clean reads | 总对比到的序列数 Total mapped reads | 多重对比的序列数 Multiple mapped reads | 唯一对比的序列数 Uniquely mapped reads |
|---|---|---|---|---|---|
| Normal A1 | 60 328 616 | 59 694 488 | 50 892 462(85.25%) | 4 145 489(6.94%) | 46 746 973(78.31%) |
| Normal A2 | 53 878 956 | 53 400 340 | 45 855 826(85.87%) | 4 209 183(7.88%) | 41 646 643(77.99%) |
| Normal A3 | 65 393 020 | 64 761 370 | 55 361 124(85.48%) | 5 082 167(7.85%) | 50 278 957(77.64%) |
| GF A1 | 48 721 914 | 48 260 760 | 41 315 919(85.61%) | 2 945 880(6.10%) | 38 370 039(79.51%) |
| GF A2 | 54 262 278 | 53 749 144 | 46 093 576(85.76%) | 3 467 323(6.45%) | 42 626 253(79.31%) |
| GF A3 | 57 431 858 | 56 874 014 | 48 683 278(85.60%) | 3 484 909(6.13%) | 45 198 369(79.47%) |
表1 测序数据基因组比对统计
Table 1 Statistical list of mapping to genome
| 样品名 Sample name | 总序列数 Raw reads | 干净序列数 Clean reads | 总对比到的序列数 Total mapped reads | 多重对比的序列数 Multiple mapped reads | 唯一对比的序列数 Uniquely mapped reads |
|---|---|---|---|---|---|
| Normal A1 | 60 328 616 | 59 694 488 | 50 892 462(85.25%) | 4 145 489(6.94%) | 46 746 973(78.31%) |
| Normal A2 | 53 878 956 | 53 400 340 | 45 855 826(85.87%) | 4 209 183(7.88%) | 41 646 643(77.99%) |
| Normal A3 | 65 393 020 | 64 761 370 | 55 361 124(85.48%) | 5 082 167(7.85%) | 50 278 957(77.64%) |
| GF A1 | 48 721 914 | 48 260 760 | 41 315 919(85.61%) | 2 945 880(6.10%) | 38 370 039(79.51%) |
| GF A2 | 54 262 278 | 53 749 144 | 46 093 576(85.76%) | 3 467 323(6.45%) | 42 626 253(79.31%) |
| GF A3 | 57 431 858 | 56 874 014 | 48 683 278(85.60%) | 3 484 909(6.13%) | 45 198 369(79.47%) |
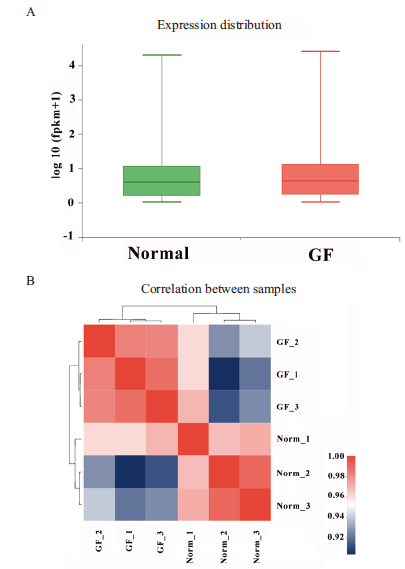
图4 样本基因表达分布和相关性 A:基因表达量分布情况;B:样本相关性
Fig.4 Distribution and correlation of genes expression patterns in different samples A:The distribution of gene expression. B:Correlation of samples

图5 差异表达基因 A:差异表达基因维恩图;B:差异基因火山图;C:差异基因聚类图
Fig.5 Differentially expressed genes A:The Veen map of differentially expressed gene. B:The volcano map of differentially expressed gene. C:The clustering map of differentially expressed gene
| 基因名称Gene Name | log2FC | 变化Change | 功能Function |
|---|---|---|---|
| TNS4 | 4.756 | Up | Function unknown |
| TMP | 4.587 | Up | Function unknown |
| NRIP3 | 4.305 | Up | Proteolysis,aspartic-type endopeptidase activity |
| MAPKAPK | 4.088 | Up | ATP binding,protein kinase activity |
| PRG4 | 4.076 | Up | Receptor-mediated endocytosis |
| IRF5 | 4.051 | Up | Function unknown |
| IRF6 | 4.007 | Up | Keratinocyte proliferation |
| ITIH3 | 3.668 | Up | Hyaluronan metabolic process |
| 3,5,3’,5’- tetraiodothyronine | 3.566 | Up | Hormone biosynthetic process |
| Phenylalanine 4-monooxygenase | 3.55 | Up | Amino acid transport and metabolism,oxidoreductase activity |
| C4BPA | -7.537 | Down | Positive regulation of protein catabolic |
| SQRDL | -6.269 | Down | Oxidoreductase activity |
| PIK3C2G | -4.847 | Down | Phosphatidylinositol 3-kinase(PI3K)complex |
| KCNIP2 | -4.293 | Down | Cell cycle control,myosin light chain |
| LIPG | -4.149 | Down | Lipid metabolic,high-density lipoprotein particle remodeling |
| MAP6 | -3.894 | Down | Lysosome localization |
| CES1 | -3.625 | Down | Lipid transport and metabolism,Carboxylesterase |
| CYP1A1 | -3.588 | Down | Steroid metabolic,Steroid hormone biosynthesis |
| SucCβ | -3.498 | Down | Fatty acid biosynthetic process,acetyl-CoA biosynthetic process |
| LRFN2 | -3.452 | Down | Function unknown |
表2 十个表达差异最显著的上调和下调基因
Table 2 Top 10 up or down expressed genes
| 基因名称Gene Name | log2FC | 变化Change | 功能Function |
|---|---|---|---|
| TNS4 | 4.756 | Up | Function unknown |
| TMP | 4.587 | Up | Function unknown |
| NRIP3 | 4.305 | Up | Proteolysis,aspartic-type endopeptidase activity |
| MAPKAPK | 4.088 | Up | ATP binding,protein kinase activity |
| PRG4 | 4.076 | Up | Receptor-mediated endocytosis |
| IRF5 | 4.051 | Up | Function unknown |
| IRF6 | 4.007 | Up | Keratinocyte proliferation |
| ITIH3 | 3.668 | Up | Hyaluronan metabolic process |
| 3,5,3’,5’- tetraiodothyronine | 3.566 | Up | Hormone biosynthetic process |
| Phenylalanine 4-monooxygenase | 3.55 | Up | Amino acid transport and metabolism,oxidoreductase activity |
| C4BPA | -7.537 | Down | Positive regulation of protein catabolic |
| SQRDL | -6.269 | Down | Oxidoreductase activity |
| PIK3C2G | -4.847 | Down | Phosphatidylinositol 3-kinase(PI3K)complex |
| KCNIP2 | -4.293 | Down | Cell cycle control,myosin light chain |
| LIPG | -4.149 | Down | Lipid metabolic,high-density lipoprotein particle remodeling |
| MAP6 | -3.894 | Down | Lysosome localization |
| CES1 | -3.625 | Down | Lipid transport and metabolism,Carboxylesterase |
| CYP1A1 | -3.588 | Down | Steroid metabolic,Steroid hormone biosynthesis |
| SucCβ | -3.498 | Down | Fatty acid biosynthetic process,acetyl-CoA biosynthetic process |
| LRFN2 | -3.452 | Down | Function unknown |
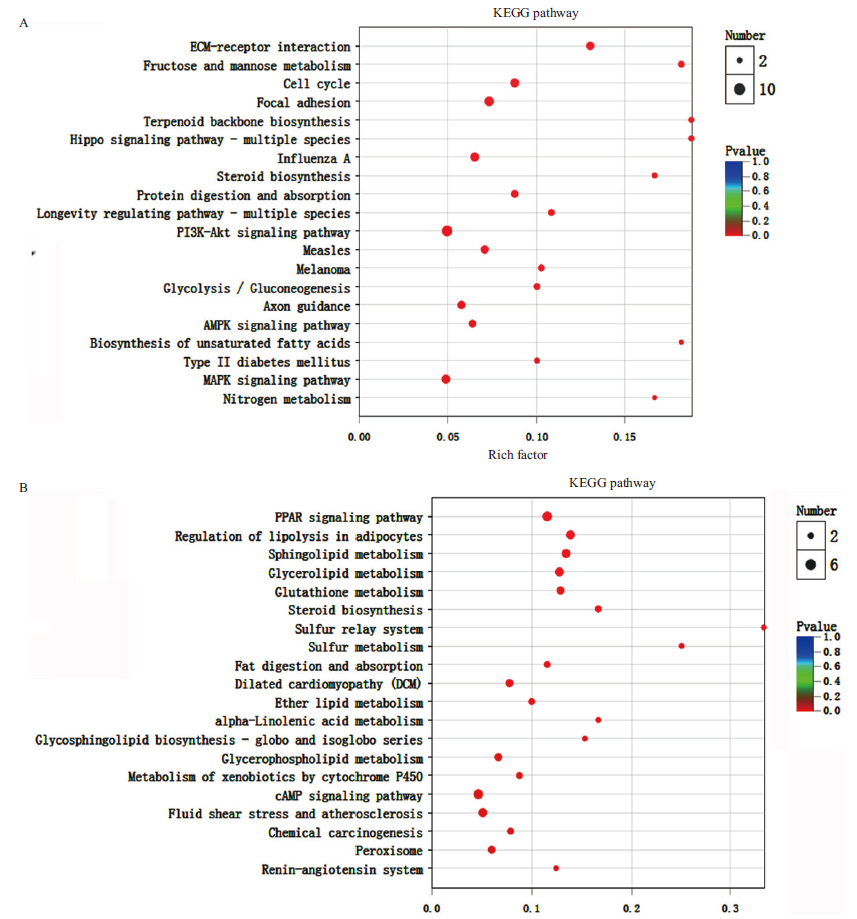
图7 差异表达基因的KEGG功能富集 A:下调基因的KEGG分析;B:上调基因的KEGG分析
Fig.7 KEGG enrichment analysis of the differentially expressed genes A: KEGG analysis of the down-regulated genes. B:KEGG analysis of the up-regulated genes
| [1] |
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome[J]. Nature, 2012,486(7402):207-214.
doi: 10.1038/nature11234 URL |
| [2] |
Kim HB, Isaacson RE. The pig gut microbial diversity:Understanding the pig gut microbial ecology through the next generation high throughput sequencing[J]. Veterinary Microbiology, 2015,177(3):242-251.
doi: 10.1016/j.vetmic.2015.03.014 URL |
| [3] | 唐义梅. 梅山猪粪便微生物移植对长×大后备母猪卵泡发育的影响[D]. 武汉:华中农业大学, 2019. |
| Tang YM. Effect of fecal microbiota transplantation of Meishan pig on follicle development in Landrace × Yorkshire gilts[D]. Wuhan:Huazhong Agricultural University, 2019. | |
| [4] |
Ejtahed HS, Hasani-Ranjbar S. Neuromodulatory effect of microbiome on gut-brain axis;new target for obesity drugs[J]. Journal of Diabetes and Metabolic Disorders, 2019,18(1):263-265.
doi: 10.1007/s40200-019-00384-4 URL |
| [5] |
Fukui H. Role of gut dysbiosis in liver diseases:what have we learned so far?[J]. Diseases, 2019,7(4):58-65.
doi: 10.3390/diseases7040058 URL |
| [6] |
Gao J, Xu K, Liu H, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism[J]. Frontiers in Cellular and Infection Microbiology, 2018,8(13):16-24.
doi: 10.3389/fcimb.2018.00016 URL |
| [7] |
Boulangé CL, Neves AL, Chilloux J, et al. Impact of the gut microbiota on inflammation, obesity, and metabolic disease[J]. Genome Medicine, 2016,8(1):42-58.
doi: 10.1186/s13073-016-0303-2 pmid: 27098727 |
| [8] | Ebrahimzadeh LH, Sanaie S, Sadeghpour HF, et al. From role of gut microbiota to microbial-based therapies in type 2-diabetes[J]. Infection, Genetics and Evolution, 2020,81(1):153-167. |
| [9] |
Miraglia F, Colla E. Microbiome, Parkinson’s disease and molecular mimicry[J]. Cells, 2019,8(3):222-230.
doi: 10.3390/cells8030222 URL |
| [10] |
Heym N, Heasman BC, Hunter K, et al. The role of microbiota and inflammation in self-judgement and empathy:implications for understanding the brain-gut-microbiome axis in depression[J]. Psychopharmacology, 2019,236(5):1459-1470.
doi: 10.1007/s00213-019-05230-2 pmid: 30955108 |
| [11] | 张学娇. YAP蛋白对脂肪功能的影响及其在肥胖相关代谢性疾病中的作用[D]. 天津:天津医科大学, 2017. |
| Zhang XJ. Role of YAP in adipose tissue function and the obesity related metabolic diseases[D]. Tianjin:Tianjin Medical University, 2017. | |
| [12] | 张震. 鼠李糖乳酸杆菌LGG来源胞外多糖抑制动物脂肪生成的研究[D]. 北京:中国农业科学院, 2017. |
| Zhang Z. Isolated exopolysaccharides from Lactobacillus rhamnosus GG inhibit adipogenesis in animals[D]. Beijing:Chinese Academy of Agricultural Sciences, 2017. | |
| [13] |
Cani PD. Metabolism in 2013:The gut microbiota manages host metabolism[J]. Nature Reviews Endocrinology, 2014,10(2):74-76.
doi: 10.1038/nrendo.2013.240 URL |
| [14] | Campbell JH, Foster CM, Vishnivetskaya T, et al. Host genetic and environmental effects on mouse intestinal microbiota[J]. Multidisciplinary Journal of Microbial Ecology, 2012,6(1):2033-2044. |
| [15] |
Dhakal S, Wang L, Antony L, et al. Amish(Rural)vs. non-Amish(Urban)infant fecal microbiotas are highly diverse and their transplantation lead to differences in mucosal immune maturation in a humanized germfree piglet model[J]. Frontiers in Immunology, 2019,10(1):1509-1517.
doi: 10.3389/fimmu.2019.01509 URL |
| [16] | Potockova H, Sinkorova J, Karova K, Sinkora M. The distribution of lymphoid cells in the small intestine of germ-free and conventional piglets[J]. Devlopmental and Comparative Immunology, 2015,51(1):99-107. |
| [17] |
Sinkora M, Butler JE, Lager KM, et al. The comparative profile of lymphoid cells and the T and B cell spectratype of germ-free piglets infected with viruses SIV, PRRSV or PCV2[J]. Veterinary Research, 2014,45(1):91-105.
doi: 10.1186/s13567-014-0091-x URL |
| [18] | Xiao L, Estellé J, Kiilerich P, et al. A reference gene catalogue of the pig gut microbiome[J]. Nature Microbiology, 2016,1(1):179-184. |
| [19] | 孙静, 杜蕾, 丁玉春, 等. 无菌猪的制备与微生物质量控制[J]. 中国实验动物学报, 2017,25(6):699-702. |
| Sun J, Du L, Ding YC, et al. Breeding and microbiological quality control of germ-free pigs[J]. Acta Laboratorium Animalis Scientia Sinica, 2017,25(6):699-702. | |
| [20] |
Hu J, Lin S, Zheng B, et al. Short-chain fatty acids in control of energy metabolism[J]. Critical Reviews in Food Science and Nutrition, 2018,58(8):1243-1249.
doi: 10.1080/10408398.2016.1245650 URL |
| [21] |
Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. PNAS, 2004,101(44):15718-15723.
doi: 10.1073/pnas.0407076101 URL |
| [22] |
Tilg H, Moschen AR. Microbiota and diabetes:an evolving relationship[J]. Gut, 2014,63(9):1513-1521.
doi: 10.1136/gutjnl-2014-306928 URL |
| [23] |
Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins[J]. Nature, 2009,457(28):480-484.
doi: 10.1038/nature07540 URL |
| [24] | Toda G, Soeda K, Okazaki Y, et al. Insulin- and lipopolysaccharide-mediated signaling in adipose tissue macrophages regulates postprandial glycemia through Akt-mTOR activation[J]. Moleculer Cell, 2020,79(1):43-53. |
| [25] |
Ding XM, Li DD, Bai SP, et al. Effect of dietary xylooligosaccharides on intestinal characteristics, gut microbiota, cecal short-chain fatty acids, and plasma immune parameters of laying hens[J]. Poultry Science, 2018,97(3):874-881.
doi: 10.3382/ps/pex372 pmid: 29294100 |
| [26] | Massier L, Chakaroun R, Tabei S, et al. Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes[J]. Gut, 2020,4(2):21-41. |
| [27] |
Russo R, Cristiano C, Avagliano C, et al. Gut-brain axis:role of lipids in the regulation of inflammation, pain and CNS diseases[J]. Current Medicinal Chemistry, 2018,25(32):3930-3952.
doi: 10.2174/0929867324666170216113756 URL |
| [28] |
Muller PA, Schneeberger M, Matheis F, et al. Microbiota modulate sympathetic neurons via a gut-brain circuit[J]. Nature, 2020,583(7823):441-446.
doi: 10.1038/s41586-020-2474-7 URL |
| [29] |
Perry RJ, Peng L, Barry N. A, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome[J]. Nature, 2016,534(7606):213-217.
doi: 10.1038/nature18309 URL |
| [30] |
Brown AJ, Goldsworthy SM, Barnes, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids[J]. Journal of Biological Chemistry, 2003,278(13):11312-11319.
doi: 10.1074/jbc.M211609200 URL |
| [31] |
Xiong Y, Miyamoto N, Shibata K, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41[J]. PNAS, 2004,101(4):1045-1050.
doi: 10.1073/pnas.2637002100 URL |
| [32] | Gaestel M. MAPK-Activated protein kinases(MKs):novel insights and challenges[J]. Frontiers in Cell and Development Biology, 2016,3(8):88-96. |
| [33] |
Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases[J]. Microbiology and Molecular Biology Reviews, 2011,75(1):50-83.
doi: 10.1128/MMBR.00031-10 URL |
| [34] | Sidarala V, Kowluru A. The regulatory roles of Mitogen-Activated Protein Kinase(MAPK)pathways in health and diabetes:lessons learned from the pancreatic β-Cell[J]. Metabolic and Immune Drug Discovery, 2017,10(2):76-84. |
| [35] | Wickramasekara RN, Morrill S, Farhat Y, et al. Glutathione and Inter-α-trypsin inhibitor heavy chain 3(Itih3)mRNA levels in nicotine-treated Cd44 knockout mice[J]. Toxicology Reports, 2018,22(5):759-764. |
| [36] |
Bost F, Diarra-Mehrpour M, Martin JP. Inter-alpha-trypsin inhibitor proteoglycan family--a group of proteins binding and stabilizing the extracellular matrix[J]. European Journal of Biochemisty, 1998,252(3):339-346.
doi: 10.1046/j.1432-1327.1998.2520339.x URL |
| [37] |
Lichter-Konecki U, Vockley J. Phenylketonuria:current treatments and future developments[J]. Drugs, 2019,79(5):495-500.
doi: 10.1007/s40265-019-01079-z pmid: 30864096 |
| [38] |
Braccini L, Ciraolo E, Campa CC, et al. PI3K-C2γ is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling[J]. Nature Communications, 2015,6(1):7400-7415.
doi: 10.1038/ncomms8400 URL |
| [39] | Yu JE, Han SY, Wolfson B, et al. The role of endothelial lipase in lipid metabolism, inflammation, and cancer[J]. Histology and Histopatholoy, 2018,33(1):1-10. |
| [40] |
Lian J, Nelson R, Lehner R. Carboxylesterases in lipid metaboli-sm:from mouse to human[J]. Protein Cell, 2018,9(1):178-195.
doi: 10.1007/s13238-017-0437-z URL |
| [41] | Wang D, Zou L, Jin Q, et al. Human carboxylesterases:a compr-ehensive review[J]. Acta Pharm Sin B, 2018,8(5):9-22. |
| [42] | Zhang WY, Wang H, Qi S, et al. CYP1A1 relieves lipopolysacc-haride-induced inflammatory responses in Bovine mammary epithelial cells[J]. Mediators Inflamm, 2018,1(3):1-10. |
| [43] |
Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. PNAS, 2004,101(44):15718-15723.
doi: 10.1073/pnas.0407076101 URL |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [3] | 刘玉玲, 王梦瑶, 孙琦, 马利花, 朱新霞. 启动子RD29A对转雪莲SikCDPK1基因烟草抗逆性的影响[J]. 生物技术通报, 2023, 39(9): 168-175. |
| [4] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [5] | 王贵芳, 姚元涛, 许海峰, 相昆, 梁家慧, 张淑辉, 王文茹, 张明娟, 张美勇, 陈新. 核桃JrSnRK1α1.1调控种子油脂合成与积累[J]. 生物技术通报, 2023, 39(9): 183-191. |
| [6] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [7] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [8] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [9] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [10] | 陈金行, 张逸, 张军涛, 未本美, 王宏勋, 郑明明. 固定化脂肪酶的创制及其在乙酸肉桂酯无溶剂制备中的应用[J]. 生物技术通报, 2023, 39(9): 97-104. |
| [11] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [12] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [13] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [14] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [15] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||