








生物技术通报 ›› 2023, Vol. 39 ›› Issue (9): 105-116.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0091
林红妍1,2,3( ), 郭晓蕊1,2,3, 刘迪1,2,3, 李慧1,2,3, 陆海1,2,3(
), 郭晓蕊1,2,3, 刘迪1,2,3, 李慧1,2,3, 陆海1,2,3( )
)
收稿日期:2023-02-07
出版日期:2023-09-26
发布日期:2023-10-24
通讯作者:
陆海,男,博士,教授,研究方向:树木分子生物学;E-mail: luhai1974@bjfu.edu.cn作者简介:林红妍,女,硕士研究生,研究方向:树木分子生物学;E-mail: 2638859438@qq.com
基金资助:
LIN Hong-yan1,2,3( ), GUO Xiao-rui1,2,3, LIU Di1,2,3, LI Hui1,2,3, LU Hai1,2,3(
), GUO Xiao-rui1,2,3, LIU Di1,2,3, LI Hui1,2,3, LU Hai1,2,3( )
)
Received:2023-02-07
Published:2023-09-26
Online:2023-10-24
摘要:
细胞壁是植物细胞特有的结构,在参与形态建成、水分和营养物质运输以及抵御生物和非生物胁迫中发挥重要作用。前人研究表明AtbHLH68作为bHLH蛋白的第10亚家族成员,在拟南芥茎维管组织中表达。为探究其在细胞壁发育方面的分子机制,本研究建立了雌二醇诱导pER8-AtbHLH68拟南芥表达系统,实时荧光定量PCR(RT-qPCR)结果显示,10 μmol/L雌二醇处理4 h能够有效诱导AtbHLH68的表达,并且随着诱导时间的增加AtbHLH68的mRNA水平进一步积累,处理8 h后达到对照组29倍。利用转录组测序分析得到了在拟南芥茎中AtbHLH68诱导表达后产生的差异基因。与对照组相比,10 μmol/L雌二醇处理8 h后,共得到差异基因2 334个,其中831个基因显著上调,1 503个基因显著下调。显著富集的GO词条主要与细胞壁组分、防御响应、果胶修饰与降解以及激素响应有关。KEGG分析表明DEGs参与了果胶修饰或木质素生物合成代谢等过程。这些结果表明参与以上过程的差异基因可能被AtbHLH68直接或间接调控,从而导致拟南芥茎细胞壁组分相关基因表达量的变化。该研究为阐明转录因子AtbHLH68在细胞壁发育中的分子机制提供了理论依据。
林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116.
LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis[J]. Biotechnology Bulletin, 2023, 39(9): 105-116.
| 基因ID Unigene ID | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| AT3G18780 | Actin2-F | CGTATGAGCAAGGAGATCAC |
| Actin2-R | CACATCTGTTGGAAGGTGCT | |
| AT4G29100 | bHLH68-F | ACCACGGATGATGCCCACT |
| bHLH68-R | CCATCATCAATCCACCCAAAA | |
| AT1G56100 | PMEI-F | TGGTTGCCTCGTTGGATG |
| PMEI-R | TGTGTAGGTTGACCATTGATGTAAA | |
| AT2G47030 | VGD1-F | GCACAGTCCAGGTTGAGTCCGAAGG |
| VGD1-R | GGTCTCCGTTCACTCTGATAGCCAC | |
| AT4G34230 | CAD5-F | GTTGCCTCGTTGGATGTTGC |
| CAD5-R | CGCCTTGTGTAGGTTGACCATTGAT | |
| AT5G62350 | PMEI-F | ACTAAGCCGAGCCCAATCC |
| PMEI-R | GTATCGTTCATCTCCTCGACGCAAT | |
| AT3G56400 | WRKY70-F | GCGATAGTCGGAAAAGATTGG |
| WRKY70-R | TGGGAATTTGGCATTAAGAA | |
| AT1G30760 | BBE13-F | TCGGCGTAATACTTGCCTGGAAAAT |
| BBE13-R | TATCAGCGACTTGTTGCCACTT | |
| YXM-pER8-bHLH68-F | GGTAATGCCATGTAATATGCTCG | |
| YXM-pER8-bHLH68-R | CATCATCAATCCACCCAAAA |
表1 引物序列
Table1 Primer sequence
| 基因ID Unigene ID | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| AT3G18780 | Actin2-F | CGTATGAGCAAGGAGATCAC |
| Actin2-R | CACATCTGTTGGAAGGTGCT | |
| AT4G29100 | bHLH68-F | ACCACGGATGATGCCCACT |
| bHLH68-R | CCATCATCAATCCACCCAAAA | |
| AT1G56100 | PMEI-F | TGGTTGCCTCGTTGGATG |
| PMEI-R | TGTGTAGGTTGACCATTGATGTAAA | |
| AT2G47030 | VGD1-F | GCACAGTCCAGGTTGAGTCCGAAGG |
| VGD1-R | GGTCTCCGTTCACTCTGATAGCCAC | |
| AT4G34230 | CAD5-F | GTTGCCTCGTTGGATGTTGC |
| CAD5-R | CGCCTTGTGTAGGTTGACCATTGAT | |
| AT5G62350 | PMEI-F | ACTAAGCCGAGCCCAATCC |
| PMEI-R | GTATCGTTCATCTCCTCGACGCAAT | |
| AT3G56400 | WRKY70-F | GCGATAGTCGGAAAAGATTGG |
| WRKY70-R | TGGGAATTTGGCATTAAGAA | |
| AT1G30760 | BBE13-F | TCGGCGTAATACTTGCCTGGAAAAT |
| BBE13-R | TATCAGCGACTTGTTGCCACTT | |
| YXM-pER8-bHLH68-F | GGTAATGCCATGTAATATGCTCG | |
| YXM-pER8-bHLH68-R | CATCATCAATCCACCCAAAA |
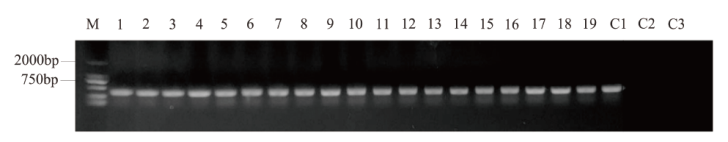
图1 PCR法筛选转基因pER8-AtbHLH68拟南芥阳性苗 M:2 000 bp marker;1-19:pER8-AtbHLH68转基因阳性植株;C1:质粒对照;C2:野生型拟南芥对照;C3:阴性对照
Fig. 1 Screening of transgenic pER8-AtbHLH68 Arabidop-sis positive plant by PCR M:2 000 bp marker;1-19: pER8-AtbHLH68 transgenic positive plants. C1: Plasmid control. C2: Wild-type Arabidopsis. C3: Negative control
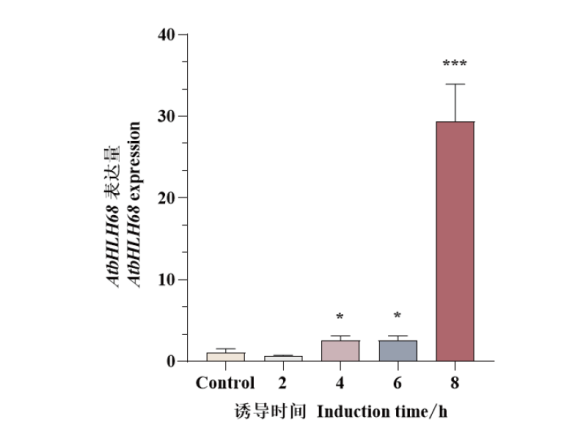
图2 RT-qPCR分析不同时长雌二醇处理下AtbHLH68基因表达量 Control:DMSO溶液处理8 h;2 h、4 h、6 h、8 h:雌二醇溶液处理拟南芥茎2 h、4 h、6 h、8 h;* P<0.05;** P<0.01;*** P<0.001,下同
Fig. 2 Expression level of AtbHLH68 under estradiol treatments at different time points by RT-qPCR Control: DMSO treatment for 8 h. 2 h, 4 h, 6 h, 8 h: 10 μmol/L estradiol treatment of Arabidopsis stem for 2 h, 4 h, 6 h, 8 h. * P<0.05. ** P<0.01. *** P<0.001. The same below
| 样品名 Sample | 原始序列数 Raw reads/M | 高质量序列数 Clean reads/M | Q20/% | Q30/% | 比对到参考基因组序列数 No. of alignment reference genome sequences/% | 唯一参考序列数 No. of unique alignment sequences/% |
|---|---|---|---|---|---|---|
| 4 h | 47.33 | 45.22 | 98.02 | 94.51 | 96.83 | 95.08 |
| 6 h | 45.57 | 43.95 | 97.83 | 93.99 | 97.12 | 95.35 |
| 8 h | 45.57 | 44.01 | 97.78 | 93.86 | 96.74 | 94.97 |
| Control | 47.33 | 45 | 97.9 | 94.18 | 96.13 | 94.11 |
表2 转录组测序质控数据
Table 2 Quality control data for transcriptome sequencing
| 样品名 Sample | 原始序列数 Raw reads/M | 高质量序列数 Clean reads/M | Q20/% | Q30/% | 比对到参考基因组序列数 No. of alignment reference genome sequences/% | 唯一参考序列数 No. of unique alignment sequences/% |
|---|---|---|---|---|---|---|
| 4 h | 47.33 | 45.22 | 98.02 | 94.51 | 96.83 | 95.08 |
| 6 h | 45.57 | 43.95 | 97.83 | 93.99 | 97.12 | 95.35 |
| 8 h | 45.57 | 44.01 | 97.78 | 93.86 | 96.74 | 94.97 |
| Control | 47.33 | 45 | 97.9 | 94.18 | 96.13 | 94.11 |
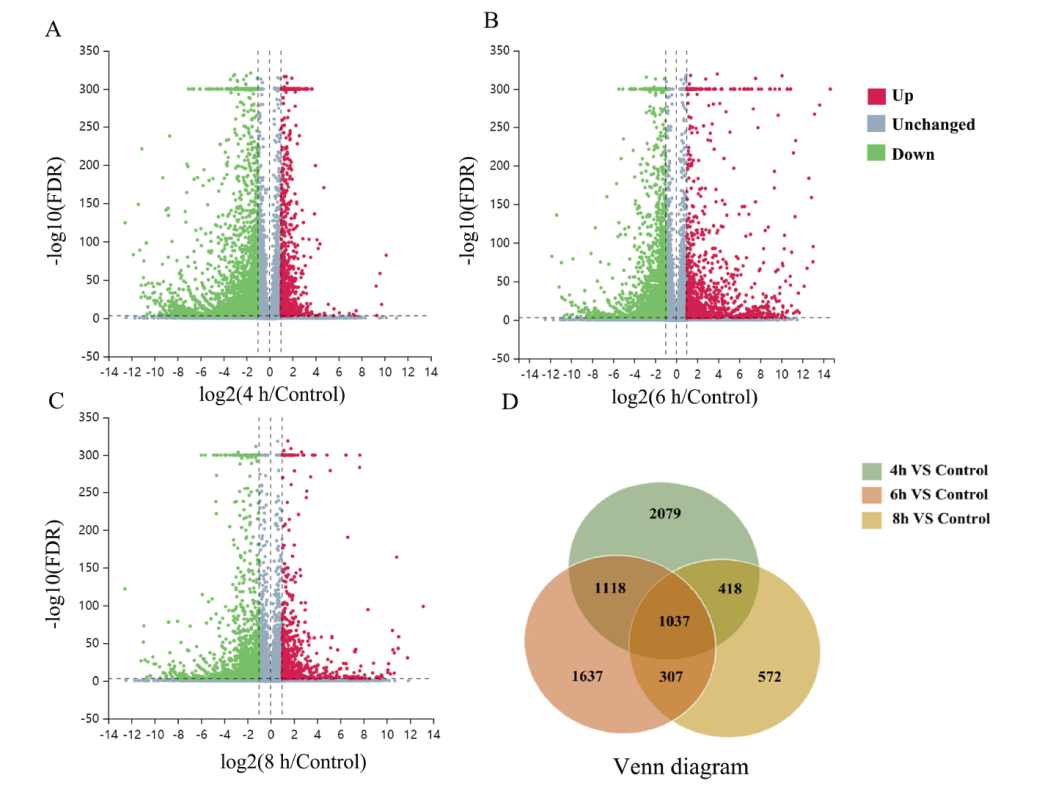
图3 4 h、6 h、8 h差异基因数目统计和韦恩图 A:4 h差异基因火山图;B:6 h 差异基因火山图;C:8 h 差异基因火山图;D:4 h、6 h、8 h差异基因韦恩图;红色圆点:上调差异基因;绿色圆点:下调差异基因;蓝色圆点:无差异基因
Fig. 3 4 h, 6 h and 8 h DEGs statistical number and Venn diagram A: 4 h volcano map of DEGs. B: 6 h volcano map of DEGs. C: 8 h volcano map of DEGs. D: 4 h, 6 h, 8 h Venn diagram of DGEs. Red dots: Up-regulated DEGs. Green dots: Down-regulated DEGs. Blue dots: Genes expressed without significant difference
| 基因表达Gene expression | 基因号Gene ID | Log2x | P值P Value | 描述Description |
|---|---|---|---|---|
| 下调基因 | AT2G06850 | -1.06394221 | 1.85E-50 | Xyloglucan endotransglucosylase/hydrolase 4 |
| Down-regulated gene | AT2G36870 | -1.266714752 | 3.34E-19 | Xyloglucan endotransglucosylase/hydrolase 32 |
| AT4G25820 | -2.74819285 | 1.28E-06 | Xyloglucan endotransglucosylase/hydrolase 14 | |
| AT5G57530 | -5.654911378 | 8.25E-28 | Xyloglucan endotransglucosylase/hydrolase 12 | |
| AT5G57540 | -3.641717279 | 7.07E-17 | Xyloglucan endotransglucosylase/hydrolase 13 | |
| AT1G71695 | -1.298988679 | 3.66E-22 | Peroxidase superfamily protein | |
| AT1G30870 | -3.50779464 | 6.16E-08 | Peroxidase 7 | |
| AT3G49110 | -2.420843121 | 6.80E-11 | Peroxidase 33 | |
| AT3G50990 | -2.306246473 | 8.33E-06 | Peroxidase 36 | |
| AT4G37980 | -1.756033681 | 3.73E-18 | Cinnamyl-alcohol dehydrogenase 7 | |
| AT2G47030 | -2.29336778 | 2.45E-142 | Pectin methylesterase 4 | |
| AT2G47040 | -2.786313785 | 0 | Vanguard1, VGD1 | |
| AT3G10720 | -2.090197809 | 2.92E-13 | Pectin methylesterase 25 | |
| AT3G62170 | -3.528498693 | 4.35E-207 | Pectin methylesterase 37 | |
| AT4G15980 | -2.788495895 | 1.82E-05 | Pectin methylesterase 43 | |
| AT5G07410 | -4.717215431 | 2.64E-240 | Pectin methylesterase 48 | |
| AT3G05610 | -2.626956589 | 1.03E-120 | Pectin methylesterase 21 | |
| AT2G26440 | -2.370550991 | 3.78E-132 | Pectin methylesterase 12 | |
| AT3G06830 | -2.258402957 | 2.42E-15 | Plant invertase/pectin methylesterase inhibitor superfamily | |
| AT5G07420 | -4.837943242 | 2.77E-08 | Pectin lyase-like superfamily protein | |
| AT5G07430 | -2.606610505 | 7.12E-169 | Pectin lyase-like superfamily protein | |
| AT3G17060 | -2.165538573 | 6.39E-22 | Pectin lyase-like superfamily protein | |
| AT1G14420 | -2.298373367 | 3.91E-46 | Pectin lyase-like superfamily protein | |
| AT2G02720 | -2.19730144 | 4.49E-24 | Pectin lyase-like superfamily protein | |
| AT3G01270 | -2.71241092 | 0 | Pectin lyase-like superfamily protein | |
| AT5G15110 | -2.336194664 | 2.35E-29 | Pectate lyase family protein | |
| AT5G48900 | -6.312882955 | 1.10E-22 | Pectin lyase-like superfamily protein | |
| AT5G55720 | -3.135159583 | 4.76E-11 | Pectin lyase-like superfamily protein | |
| AT3G45860 | -1.803254412 | 3.34E-18 | Cysteine-rich RLK(receptor-like protein kinase)4 | |
| AT4G23170 | -1.533408039 | 1.62E-109 | Cysteine-rich RLK(receptor-like protein kinase)9 | |
| AT2G40750 | -1.152515323 | 8.63E-64 | WRKY DNA-binding protein 54 | |
| AT2G46400 | -1.299919474 | 3.63E-61 | WRKY DNA-binding protein 46 | |
| AT3G56400 | -1.40629482 | 0 | WRKY DNA-binding protein 70 | |
| AT4G23810 | -1.452162691 | 1.12E-65 | WRKY DNA-binding protein 53 | |
| AT5G24780 | -1.294335074 | 0 | Vegetative storage protein 1 | |
| AT3G16470 | -1.212665758 | 0 | A JA-responsive gene that coordinates with GRP 7 | |
| AT2G32690 | -2.181267688 | 6.01E-32 | Glycine-rich protein 23 | |
| AT1G20510 | -1.035196623 | 4.70E-93 | OPC-8:CoA ligase involved in jasmonic acid biosynthesis | |
| 上调基因 | AT1G11545 | 2.054076673 | 1.34E-06 | Xyloglucan endotransglucosylase/hydrolase 8 |
| Up-regulated gene | AT3G48580 | 3.262777493 | 3.34E-61 | Xyloglucan endotransglucosylase/hydrolase 11 |
| AT4G14130 | 1.838579331 | 4.09E-18 | Xyloglucan endotransglucosylase/hydrolase 15 | |
| AT4G37800 | 2.408582477 | 8.79E-21 | Xyloglucan endotransglucosylase/hydrolase 7 | |
| AT1G30700 | 3.103198888 | 3.07E-254 | FAD-binding berberine family protein, AtBBE 7 | |
| AT1G30720 | 3.04026387 | 2.09E-22 | FAD-binding berberine family protein, AtBBE 10 | |
| AT1G30730 | 2.767165832 | 8.28E-09 | FAD-binding berberine family protein, AtBBE 11 | |
| AT4G20800 | 3.940837928 | 8.47E-14 | FAD-binding berberine family protein, AtBBE 17 | |
| AT5G44390 | 2.190683562 | 4.52E-05 | FAD-binding berberine family protein, AtBBE 25 | |
| AT5G44400 | 2.779652634 | 0 | FAD-binding berberine family protein, AtBBE 26 | |
| AT1G30760 | 1.250535773 | 3.26e-210 | Berberine bridge enzyme-like 13, AtBBE 13 | |
| AT2G34790 | 2.036462643 | 1.06E-182 | Berberine bridge enzyme-like 15, AtBBE 15 | |
| AT4G36430 | 2.677849798 | 3.42E-36 | Peroxidase superfamily protein | |
| AT3G21770 | 1.766366511 | 5.33E-07 | Peroxidase superfamily protein | |
| AT3G28200 | 1.80094768 | 1.06E-34 | Peroxidase superfamily protein | |
| AT1G05260 | 1.600816452 | 2.62E-31 | Peroxidase 3 | |
| AT5G05340 | 2.40141963 | 1.48E-80 | Peroxidase 52 | |
| AT5G60020 | 1.132349101 | 5.96E-135 | Laccase 17 | |
| AT4G34230 | 1.168213629 | 0 | Cinnamyl alcohol dehydrogenase 5 | |
| AT4G37990 | 1.401205226 | 3.90E-06 | Cinnamyl-alcohol dehydrogenase 8 | |
| AT1G11590 | 2.950611973 | 1.48E-19 | Pectin methylesterase 19 | |
| AT5G09760 | 1.08363098 | 2.08E-22 | Pectin methylesterase 15 | |
| AT1G14890 | 1.984238146 | 0 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT5G62350 | 1.901654358 | 0 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT1G62770 | 1.620506618 | 3.20E-05 | Pectin methyleseterase inhibitor 19 | |
| AT1G56100 | 1.25 | 0 | Plant invertase/pectin methylesterase inhibitor 14 | |
| AT3G47380 | 1.787395021 | 2.75E-23 | Plant invertase/pectin methylesterase inhibitor 11 | |
| AT3G27400 | 2.985665084 | 5.10E-106 | Pectin lyase-like superfamily protein | |
| AT3G53190 | 1.145912382 | 1.86E-14 | Pectin lyase-like superfamily protein | |
| AT4G13710 | 1.186011986 | 2.50E-09 | Pectin lyase-like superfamily protein | |
| AT4G24780 | 1.1622914 | 1.26E-84 | Pectate lyase 19 | |
| AT5G04310 | 1.318331844 | 6.35E-11 | Pectate lyase like 12 |
表3 部分差异基因功能分析
Table 3 Functional analysis of DEGs
| 基因表达Gene expression | 基因号Gene ID | Log2x | P值P Value | 描述Description |
|---|---|---|---|---|
| 下调基因 | AT2G06850 | -1.06394221 | 1.85E-50 | Xyloglucan endotransglucosylase/hydrolase 4 |
| Down-regulated gene | AT2G36870 | -1.266714752 | 3.34E-19 | Xyloglucan endotransglucosylase/hydrolase 32 |
| AT4G25820 | -2.74819285 | 1.28E-06 | Xyloglucan endotransglucosylase/hydrolase 14 | |
| AT5G57530 | -5.654911378 | 8.25E-28 | Xyloglucan endotransglucosylase/hydrolase 12 | |
| AT5G57540 | -3.641717279 | 7.07E-17 | Xyloglucan endotransglucosylase/hydrolase 13 | |
| AT1G71695 | -1.298988679 | 3.66E-22 | Peroxidase superfamily protein | |
| AT1G30870 | -3.50779464 | 6.16E-08 | Peroxidase 7 | |
| AT3G49110 | -2.420843121 | 6.80E-11 | Peroxidase 33 | |
| AT3G50990 | -2.306246473 | 8.33E-06 | Peroxidase 36 | |
| AT4G37980 | -1.756033681 | 3.73E-18 | Cinnamyl-alcohol dehydrogenase 7 | |
| AT2G47030 | -2.29336778 | 2.45E-142 | Pectin methylesterase 4 | |
| AT2G47040 | -2.786313785 | 0 | Vanguard1, VGD1 | |
| AT3G10720 | -2.090197809 | 2.92E-13 | Pectin methylesterase 25 | |
| AT3G62170 | -3.528498693 | 4.35E-207 | Pectin methylesterase 37 | |
| AT4G15980 | -2.788495895 | 1.82E-05 | Pectin methylesterase 43 | |
| AT5G07410 | -4.717215431 | 2.64E-240 | Pectin methylesterase 48 | |
| AT3G05610 | -2.626956589 | 1.03E-120 | Pectin methylesterase 21 | |
| AT2G26440 | -2.370550991 | 3.78E-132 | Pectin methylesterase 12 | |
| AT3G06830 | -2.258402957 | 2.42E-15 | Plant invertase/pectin methylesterase inhibitor superfamily | |
| AT5G07420 | -4.837943242 | 2.77E-08 | Pectin lyase-like superfamily protein | |
| AT5G07430 | -2.606610505 | 7.12E-169 | Pectin lyase-like superfamily protein | |
| AT3G17060 | -2.165538573 | 6.39E-22 | Pectin lyase-like superfamily protein | |
| AT1G14420 | -2.298373367 | 3.91E-46 | Pectin lyase-like superfamily protein | |
| AT2G02720 | -2.19730144 | 4.49E-24 | Pectin lyase-like superfamily protein | |
| AT3G01270 | -2.71241092 | 0 | Pectin lyase-like superfamily protein | |
| AT5G15110 | -2.336194664 | 2.35E-29 | Pectate lyase family protein | |
| AT5G48900 | -6.312882955 | 1.10E-22 | Pectin lyase-like superfamily protein | |
| AT5G55720 | -3.135159583 | 4.76E-11 | Pectin lyase-like superfamily protein | |
| AT3G45860 | -1.803254412 | 3.34E-18 | Cysteine-rich RLK(receptor-like protein kinase)4 | |
| AT4G23170 | -1.533408039 | 1.62E-109 | Cysteine-rich RLK(receptor-like protein kinase)9 | |
| AT2G40750 | -1.152515323 | 8.63E-64 | WRKY DNA-binding protein 54 | |
| AT2G46400 | -1.299919474 | 3.63E-61 | WRKY DNA-binding protein 46 | |
| AT3G56400 | -1.40629482 | 0 | WRKY DNA-binding protein 70 | |
| AT4G23810 | -1.452162691 | 1.12E-65 | WRKY DNA-binding protein 53 | |
| AT5G24780 | -1.294335074 | 0 | Vegetative storage protein 1 | |
| AT3G16470 | -1.212665758 | 0 | A JA-responsive gene that coordinates with GRP 7 | |
| AT2G32690 | -2.181267688 | 6.01E-32 | Glycine-rich protein 23 | |
| AT1G20510 | -1.035196623 | 4.70E-93 | OPC-8:CoA ligase involved in jasmonic acid biosynthesis | |
| 上调基因 | AT1G11545 | 2.054076673 | 1.34E-06 | Xyloglucan endotransglucosylase/hydrolase 8 |
| Up-regulated gene | AT3G48580 | 3.262777493 | 3.34E-61 | Xyloglucan endotransglucosylase/hydrolase 11 |
| AT4G14130 | 1.838579331 | 4.09E-18 | Xyloglucan endotransglucosylase/hydrolase 15 | |
| AT4G37800 | 2.408582477 | 8.79E-21 | Xyloglucan endotransglucosylase/hydrolase 7 | |
| AT1G30700 | 3.103198888 | 3.07E-254 | FAD-binding berberine family protein, AtBBE 7 | |
| AT1G30720 | 3.04026387 | 2.09E-22 | FAD-binding berberine family protein, AtBBE 10 | |
| AT1G30730 | 2.767165832 | 8.28E-09 | FAD-binding berberine family protein, AtBBE 11 | |
| AT4G20800 | 3.940837928 | 8.47E-14 | FAD-binding berberine family protein, AtBBE 17 | |
| AT5G44390 | 2.190683562 | 4.52E-05 | FAD-binding berberine family protein, AtBBE 25 | |
| AT5G44400 | 2.779652634 | 0 | FAD-binding berberine family protein, AtBBE 26 | |
| AT1G30760 | 1.250535773 | 3.26e-210 | Berberine bridge enzyme-like 13, AtBBE 13 | |
| AT2G34790 | 2.036462643 | 1.06E-182 | Berberine bridge enzyme-like 15, AtBBE 15 | |
| AT4G36430 | 2.677849798 | 3.42E-36 | Peroxidase superfamily protein | |
| AT3G21770 | 1.766366511 | 5.33E-07 | Peroxidase superfamily protein | |
| AT3G28200 | 1.80094768 | 1.06E-34 | Peroxidase superfamily protein | |
| AT1G05260 | 1.600816452 | 2.62E-31 | Peroxidase 3 | |
| AT5G05340 | 2.40141963 | 1.48E-80 | Peroxidase 52 | |
| AT5G60020 | 1.132349101 | 5.96E-135 | Laccase 17 | |
| AT4G34230 | 1.168213629 | 0 | Cinnamyl alcohol dehydrogenase 5 | |
| AT4G37990 | 1.401205226 | 3.90E-06 | Cinnamyl-alcohol dehydrogenase 8 | |
| AT1G11590 | 2.950611973 | 1.48E-19 | Pectin methylesterase 19 | |
| AT5G09760 | 1.08363098 | 2.08E-22 | Pectin methylesterase 15 | |
| AT1G14890 | 1.984238146 | 0 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT5G62350 | 1.901654358 | 0 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT1G62770 | 1.620506618 | 3.20E-05 | Pectin methyleseterase inhibitor 19 | |
| AT1G56100 | 1.25 | 0 | Plant invertase/pectin methylesterase inhibitor 14 | |
| AT3G47380 | 1.787395021 | 2.75E-23 | Plant invertase/pectin methylesterase inhibitor 11 | |
| AT3G27400 | 2.985665084 | 5.10E-106 | Pectin lyase-like superfamily protein | |
| AT3G53190 | 1.145912382 | 1.86E-14 | Pectin lyase-like superfamily protein | |
| AT4G13710 | 1.186011986 | 2.50E-09 | Pectin lyase-like superfamily protein | |
| AT4G24780 | 1.1622914 | 1.26E-84 | Pectate lyase 19 | |
| AT5G04310 | 1.318331844 | 6.35E-11 | Pectate lyase like 12 |

图6 生物合成通路图 A:戊糖葡萄糖醛酸转化通路图和差异基因热图,值为log2(FPKM+1);PME:果胶甲酯酶;PMEI果胶甲酯酶抑制剂;PG:多聚半乳糖醛酸酶;PL:多聚半乳糖醛酸裂解酶;B:苯丙烷生物合成途径通路图和差异基因热图,值为log2(FPKM+1);CAD:肉桂醇脱氢酶;PER:过氧化物酶;LAC:漆酶
Fig. 6 Biosynthetic pathway map A: Pentose and glucuronate interconversions pathway map and DEG heat map, values are log2(FPKM+1). PME: Pectin methylesterase. PMEI: Pectin methylesterase inhibitor. PG: Polygalacturonase. PL: Pectate lyase. B: Phenylpropane biosynthesis pathway pathway map and DEG heat map, values are log2(FPKM+1). CAD: Cinnamyl alcohol dehydrogenase. PER: Peroxidase. LAC: Laccase
| [1] |
Zhang BC, Gao YH, Zhang LJ, et al. The plant cell wall: Biosynthesis, construction, and functions[J]. J Integr Plant Biol, 2021, 63(1): 251-272.
doi: 10.1111/jipb.13055 |
| [2] | 张保才, 周奕华. 植物细胞壁形成机制的新进展[J]. 中国科学: 生命科学, 2015, 45(6): 544-556. |
|
Zhang BC, Zhou Y. Plant cell wall formation and regulation[J]. Sci Sin Vitae, 2015, 45(6): 544-556.
doi: 10.1360/N052015-00076 URL |
|
| [3] |
Kumar M, Atanassov I, Turner S. Functional analysis of cellulose synthase(CESA)protein class specificity[J]. Plant Physiol, 2017, 173(2): 970-983.
doi: 10.1104/pp.16.01642 URL |
| [4] |
Taylor NG. Cellulose biosynthesis and deposition in higher plants[J]. New Phytol, 2008, 178(2): 239-252.
doi: 10.1111/j.1469-8137.2008.02385.x pmid: 18298430 |
| [5] |
Scheller HV, Ulvskov P. Hemicelluloses[J]. Annu Rev Plant Biol, 2010, 61: 263-289.
doi: 10.1146/annurev-arplant-042809-112315 pmid: 20192742 |
| [6] | 陈思慧, 徐华, 黄非, 等. 草本植物半纤维素研究进展[J]. 草业科学, 2020, 37(3): 506-513. |
| Chen SH, Xu H, Huang F, et al. Progress in research of grass hemicellulose[J]. Pratacultural Sci, 2020, 37(3): 506-513. | |
| [7] |
Vanholme R, Morreel K, Darrah C, et al. Metabolic engineering of novel lignin in biomass crops[J]. New Phytol, 2012, 196(4): 978-1000.
doi: 10.1111/j.1469-8137.2012.04337.x pmid: 23035778 |
| [8] | 熊媛, 任襄襄, 朱慧, 等. 果胶甲酯酶抑制蛋白(PMEIs)的功能性研究进展[J]. 中国生物工程杂志, 2022, 42(8): 99-108. |
| Xiong Y, Ren XX, Zhu H, et al. Recent progress in the functional research of PMEIs[J]. China Biotechnol, 2022, 42(8): 99-108. | |
| [9] |
Willats WGT, Orfila C, Limberg G, et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls[J]. J Biol Chem, 2001, 276(22): 19404-19413.
doi: 10.1074/jbc.M011242200 pmid: 11278866 |
| [10] | 张海燕, 吴天祥. 微生物果胶酶的研究进展[J]. 酿酒科技, 2006(9): 82-85. |
| Zhang HY, Wu TX. Research progress in pectinase[J]. Liquor Mak Sci Technol, 2006(9): 82-85. | |
| [11] | 陈乐天, 王慧婷, 韩靖鸾, 等. 植物果胶裂解酶的研究现状及展望[J]. 华南农业大学学报, 2019, 40(5): 71-77. |
| Chen LT, Wang HT, Han JL, et al. Research progress and perspective of plant pectin lysase[J]. J South China Agric Univ, 2019, 40(5): 71-77. | |
| [12] |
Terrett OM, Dupree P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls[J]. Curr Opin Biotechnol, 2019, 56: 97-104.
doi: 10.1016/j.copbio.2018.10.010 URL |
| [13] | 余紫苹, 彭红, 林妲, 等. 植物半纤维素结构研究进展[J]. 高分子通报, 2011(6): 48-54. |
| Yu ZP, Peng H, Lin D, et al. Research progress of plant hemicellulose structure[J]. Polym Bull, 2011(6): 48-54. | |
| [14] | 刘佩佩, 张耿, 李晓娟. 植物果胶的生物合成与功能[J]. 植物学报, 2021, 56(2): 191-200. |
| Liu PP, Zhang G, Li XJ. Biosynthesis and function of plant pectin[J]. Chin Bull Bot, 2021, 56(2): 191-200. | |
| [15] | Cosgrove DJ. Growth of the plant cell wall[J]. Nat Rev Mol Cell Biol, 2005, 6(11): 850-861. |
| [16] |
Li Z, Fernie AR, Persson S. Transition of primary to secondary cell wall synthesis[J]. Sci Bull, 2016, 61(11): 838-846.
doi: 10.1007/s11434-016-1061-7 URL |
| [17] |
Heim MA, Jakoby M, Werber M, et al. The basic Helix-loop-Helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 URL |
| [18] |
Gao ZY, Sun WJ, Wang J, et al. GhbHLH18 negatively regulates fiber strength and length by enhancing lignin biosynthesis in cotton fibers[J]. Plant Sci, 2019, 286: 7-16.
doi: S0168-9452(19)30265-1 pmid: 31300144 |
| [19] |
Zhao WQ, Ding L, Liu JY, et al. Regulation of lignin biosynthesis by an atypical bHLH protein CmHLB in Chrysanthemum[J]. J Exp Bot, 2022, 73(8): 2403-2419.
doi: 10.1093/jxb/erac015 URL |
| [20] |
Yan L, Xu CH, Kang YL, et al. The heterologous expression in Arabidopsis thaliana of sorghum transcription factor SbbHLH1 downregulates lignin synthesis[J]. J Exp Bot, 2013, 64(10): 3021-3032.
doi: 10.1093/jxb/ert150 pmid: 23698629 |
| [21] |
Cao YY, Zeng HX, Ku LX, et al. ZmIBH1-1 regulates plant architecture in maize[J]. J Exp Bot, 2020, 71(10): 2943-2955.
doi: 10.1093/jxb/eraa052 pmid: 31990030 |
| [22] |
Le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana[J]. Physiol Plantarum, 2017, 160(3): 312-327.
doi: 10.1111/ppl.2017.160.issue-3 URL |
| [23] |
Ikeda M, Fujiwara S, Mitsuda N, et al. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis[J]. Plant Cell, 2012, 24(11): 4483-4497.
doi: 10.1105/tpc.112.105023 URL |
| [24] |
Oh J, Park E, Song K, et al. PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis[J]. Plant Cell, 2020, 32(1): 186-205.
doi: 10.1105/tpc.19.00515 URL |
| [25] |
Liu WW, Tai HH, Li SS, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism[J]. New Phytol, 2014, 201(4): 1192-1204.
doi: 10.1111/nph.2014.201.issue-4 URL |
| [26] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [27] |
Coculo D, Lionetti V. The plant invertase/pectin methylesterase inhibitor superfamily[J]. Front Plant Sci, 2022, 13: 863892.
doi: 10.3389/fpls.2022.863892 URL |
| [28] |
Guo XQ, Chang S, Hu JP, et al. Research progress of pectin methylesterase and its inhibitors[J]. Curr Protein Pept Sci, 2022, 23(10): 684-696.
doi: 10.2174/1389203723666220919092428 URL |
| [29] |
Shi DC, Ren AY, Tang XF, et al. MYB52 negatively regulates pectin demethylesterification in seed coat mucilage[J]. Plant Physiol, 2018, 176(4): 2737-2749.
doi: 10.1104/pp.17.01771 URL |
| [30] | Wilson MH, Holman TJ, Sørensen I, et al. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone[J]. Front Cell Dev Biol, 2015, 3: 10. |
| [31] |
Han Y, Ban QY, Hou YL, et al. Isolation and characterization of two persimmon xyloglucan endotransglycosylase/hydrolase(XTH)genes that have divergent functions in cell wall modification and fruit postharvest softening[J]. Front Plant Sci, 2016, 7: 624.
doi: 10.3389/fpls.2016.00624 pmid: 27242828 |
| [32] |
Daniel B, Pavkov-Keller T, Steiner B, et al. Oxidation of monolignols by members of the berberine bridge enzyme family suggests a role in plant cell wall metabolism[J]. J Biol Chem, 2015, 290(30): 18770-18781.
doi: 10.1074/jbc.M115.659631 pmid: 26037923 |
| [33] |
Fernández-Pérez F, Pomar F, Pedreño MA, et al. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units[J]. Physiol Plantarum, 2015, 154(3): 395-406.
doi: 10.1111/ppl.2015.154.issue-3 URL |
| [34] |
Kärkönen A, Fry SC. Effect of ascorbate and its oxidation products on H2O2 production in cell-suspension cultures of Picea abies and in the absence of cells[J]. J Exp Bot, 2006, 57(8): 1633-1644.
doi: 10.1093/jxb/erj197 pmid: 16698813 |
| [35] |
Hamann T. The plant cell wall integrity maintenance mechanism-A case study of a cell wall plasma membrane signaling network[J]. Phytochemistry, 2015, 112: 100-109.
doi: 10.1016/j.phytochem.2014.09.019 URL |
| [36] |
Zhang NL, Zhou S, Yang DY, et al. Revealing shared and distinct genes responding to JA and SA signaling in Arabidopsis by meta-analysis[J]. Front Plant Sci, 2020, 11: 908.
doi: 10.3389/fpls.2020.00908 URL |
| [37] | Chen JN, Nolan T, Ye HX, et al. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response[J]. Plant Cell, 2017: tpc. 00364.2017. |
| [1] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [2] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [3] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [4] | 张曼, 张叶卓, 何其邹洪, 鄂一岚, 李晔. 植物细胞壁结构及成像技术研究进展[J]. 生物技术通报, 2023, 39(7): 113-122. |
| [5] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [6] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [7] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [8] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [9] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [10] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制[J]. 生物技术通报, 2023, 39(11): 18-27. |
| [11] | 于波, 秦晓惠, 赵杨. 植物感应干旱信号的机制[J]. 生物技术通报, 2023, 39(11): 6-17. |
| [12] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [13] | 徐俊, 叶雨晴, 牛雅静, 黄河, 张蒙蒙. 菊花根状茎发育的转录组分析[J]. 生物技术通报, 2023, 39(10): 231-245. |
| [14] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [15] | 辛建攀, 李燕, 赵楚, 田如男. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||