








生物技术通报 ›› 2021, Vol. 37 ›› Issue (5): 273-280.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1181
瞿欢1( ), 李成1, 陈汭1, 廖艺杰1, 曹三杰1,2,3, 文翼平1, 颜其贵1, 黄小波1,2,3(
), 李成1, 陈汭1, 廖艺杰1, 曹三杰1,2,3, 文翼平1, 颜其贵1, 黄小波1,2,3( )
)
收稿日期:2020-09-18
出版日期:2021-05-26
发布日期:2021-06-11
作者简介:瞿欢,女,硕士研究生,研究方向:动物传染病;E-mail: 基金资助:
QU Huan1( ), LI Cheng1, CHEN Rui1, LIAO Yi-jie1, CAO San-jie1,2,3, WEN Yi-ping1, YAN Qi-gui1, HUANG Xiao-bo1,2,3(
), LI Cheng1, CHEN Rui1, LIAO Yi-jie1, CAO San-jie1,2,3, WEN Yi-ping1, YAN Qi-gui1, HUANG Xiao-bo1,2,3( )
)
Received:2020-09-18
Published:2021-05-26
Online:2021-06-11
摘要:
猪δ冠状病毒(Porcine deltacoronavirus,PDCoV)是一种能引起仔猪呕吐、腹泻和脱水的新的猪肠道冠状病毒,表达了PDCoV的S基因抗原表位区(S1-CTD)并建立了检测抗体的间接ELISA方法。根据S基因序列设计引物,扩增含中和抗原表位的S1-CTD区(位于S基因的832-1 848 bp),构建原核表达载体pET28a-S1-CTD,表达的重组S1-CTD蛋白经SDS-PAGE鉴定大小约41 kD,Western Blot证明其有良好的反应原性。以重组S1-CTD 蛋白为抗原建立间接ELISA,抗原最佳包被浓度为1μg/孔,待检血清的最佳稀释度为1∶50,阴阳性临界值为OD450nm≥0.377。该ELISA检测其他8种常见猪病阳性血清无交叉反应,特异性好;批内和批间变异系数均小于10%,重复性好。ELISA与病毒中和试验的总符合率达83.3%。用建立的ELISA检测了2012-2017年四川部分猪场490份临床血清,结果显示血清中PDCoV抗体阳性率为49.18%。
瞿欢, 李成, 陈汭, 廖艺杰, 曹三杰, 文翼平, 颜其贵, 黄小波. 猪δ冠状病毒S1-CTD的截短表达及间接ELISA抗体方法的建立[J]. 生物技术通报, 2021, 37(5): 273-280.
QU Huan, LI Cheng, CHEN Rui, LIAO Yi-jie, CAO San-jie, WEN Yi-ping, YAN Qi-gui, HUANG Xiao-bo. Truncated Expression of the S1-CTD Fragment of Porcine Deltacoronavirus and Establishment of an Indirect ELISA for Detecting Its Antibody[J]. Biotechnology Bulletin, 2021, 37(5): 273-280.
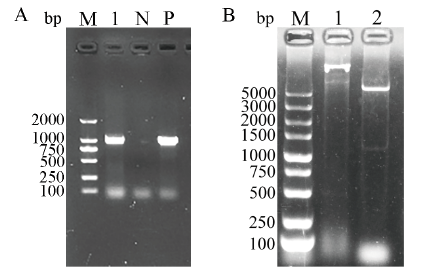
图2 重组质粒pET28a-S1CTD的PCR(A)及双酶切鉴定(B) A:重组质粒pET28a-S1CTD的PCR鉴定(M:DL2000 DNA相对分子质量标准;1:目标基因;N:阴性对照;P:阳性对照);B:重组质粒pET28a-S1CTD的双酶切鉴定(M:DL5000 DNA相对分子质量标准;1:未酶切的重组质粒;2:双酶切的重组质粒)
Fig. 2 Identification of recombinant plasmid pET28a-S1CTD by PCR (A) and double digestion (B) A: Identification of recombinant plasmid pET28a-S1CTD by PCR (M: DL2000 DNA marker, 1: Target gene, N: Negative control, P: Positive control). B: Identification of recombinant plasmid pET28a-S1CTD by double digestion (M: DL5000 DNA marke, 1: Undigested recombinant plasmid, 2: Digested recombinant plasmid)

图3 重组蛋白S1-CTD的SDS-PAGE与Western blot分析 A:重组蛋白S1-CTD的SDS-PAGE分析(M:蛋白质相对分子质量标准;1:BL21-pET28a诱导后;2:pET28a-S1-CTD诱导前;3:pET28a-S1-CTD诱导后;4:诱导后超声破碎全菌;5:超声破碎后上清液;6:超声破碎后包涵体);B:S1-CTD蛋白的纯化(M:蛋白质相对分子质量标准;1:溶解前包涵体蛋白;2: 溶解后包涵体蛋白;3:50mmol/L咪唑洗脱;4:150 mmol/L咪唑洗脱;5:250mmol/L咪唑洗脱);C:重组蛋白的Western blot分析(M:蛋白质相对分子质量标准;1-4:BL21-pET28a空载、pET28a-S1-CTD、pET-28a-E、pET-28a-N蛋白分别与猪抗PDCoV阳性血清反应;5:pET28a-S1-CTD 与猪抗PDCoV 阴性血清反应)
Fig. 3 The SDS-PAGE and Western blot analysis of recombinant S1-CTD protein A: The SDS-PAGE analysis of S1-CTD (M: Protein marker, 1: Post-induced BL21-pET28a, 2: Pre-induced pET28a-S1-CTD, 3: Post-induced pET28a-S1-CTD, 4: Hyperacoustic full bacteria after induction, 5: Hyperacoustic supernatant, 6: Hyperacoustic precipitation). B: Purification of S1-CTD protein (M: Protein marker, 1: Pre-lysing inclusion body protein, 2: Dissolved inclusion body protein, 3: Target protein eluted by 50 mmol/L imidazole, 4: Target protein eluted by 150 mmol/L imidazole, 5: Target protein eluted by 250 mmol/L imidazole). C: The western blot analysis of S1-CTD (M: Protein marker, 1-4: BL21-pET28a, pET28a-S1-CTD, pET-28a-E, pET-28a-N proteins reacted with pig anti-PDCoV positive serum, 5: pET28a-S1-CTD protein reacted with pig anti-PDCoV negative serum)
| 血清编号 Serum No. | 批内变异系数 Intra batch CV % | 批间变异系数 Inter batch CV % | ||
|---|---|---|---|---|
| ±SD | 变异系数 CV/% | ±SD | 变异系数 CV/% | |
| 1 | 0.136±0.011 | 8.07 | 0.190±0.017 | 8.85 |
| 2 | 0.110±0.011 | 9.57 | 0.217±0.020 | 9.22 |
| 3 | 0.127±0.004 | 3.48 | 0.122±0.012 | 9.98 |
| 4 | 0.124±0.005 | 4.35 | 0.155±0.013 | 8.29 |
| 5 | 0.113±0.003 | 2.65 | 0.909±0.055 | 6.01 |
| 6 | 1.251±0.043 | 3.47 | 1.022±0.079 | 7.75 |
| 7 | 0.946±0.032 | 3.43 | 0.928±0.085 | 9.12 |
表1 间接ELISA的重复性试验
Table 1 Repeatability of the indirect ELISA
| 血清编号 Serum No. | 批内变异系数 Intra batch CV % | 批间变异系数 Inter batch CV % | ||
|---|---|---|---|---|
| ±SD | 变异系数 CV/% | ±SD | 变异系数 CV/% | |
| 1 | 0.136±0.011 | 8.07 | 0.190±0.017 | 8.85 |
| 2 | 0.110±0.011 | 9.57 | 0.217±0.020 | 9.22 |
| 3 | 0.127±0.004 | 3.48 | 0.122±0.012 | 9.98 |
| 4 | 0.124±0.005 | 4.35 | 0.155±0.013 | 8.29 |
| 5 | 0.113±0.003 | 2.65 | 0.909±0.055 | 6.01 |
| 6 | 1.251±0.043 | 3.47 | 1.022±0.079 | 7.75 |
| 7 | 0.946±0.032 | 3.43 | 0.928±0.085 | 9.12 |
| 检测方法 Detection method | VNT | |||
|---|---|---|---|---|
| 阳性数 Positive number | 阴性数 Negative number | 合计 Total | ||
| 阳性数 Positive number | 9 | 3 | 12 | |
| 阴性数 Negative number | 2 | 16 | 18 | |
| 间接ELISA | 合计 Total | 11 | 19 | 30 |
| 阳性符合率 Positive coincidence rate | 81.8% | |||
| Indirect ELISA | 阴性符合率 Negative coincidence rate | 84.2% | ||
| 总符合率 Total coincidence rate | 83.3% | |||
表2 间接ELISA与VNT对血清样品的比较性检测
Table 2 Comparative detection of serum samples by indirect ELISA and VNT
| 检测方法 Detection method | VNT | |||
|---|---|---|---|---|
| 阳性数 Positive number | 阴性数 Negative number | 合计 Total | ||
| 阳性数 Positive number | 9 | 3 | 12 | |
| 阴性数 Negative number | 2 | 16 | 18 | |
| 间接ELISA | 合计 Total | 11 | 19 | 30 |
| 阳性符合率 Positive coincidence rate | 81.8% | |||
| Indirect ELISA | 阴性符合率 Negative coincidence rate | 84.2% | ||
| 总符合率 Total coincidence rate | 83.3% | |||
| 来源 Origin | 样本数 Sample number | 阳性总数 Positive number | 阳性率 Positive rate/% |
|---|---|---|---|
| 成都 Chengdu | 52 | 34 | 65.38 |
| 南充 Nanchong | 57 | 26 | 45.61 |
| 绵阳 Mianyang | 30 | 12 | 40 |
| 眉山 Meishan | 33 | 3 | 9.09 |
| 遂宁 Suining | 16 | 0 | 0 |
| 德阳 Deyang | 61 | 24 | 39.34 |
| 达州 Dazhou | 27 | 27 | 100 |
| 泸州 Luzhou | 9 | 8 | 88.89 |
| 宜宾 Yibin | 1 | 1 | 100 |
| 阿坝 Aba | 23 | 22 | 95.65 |
| 巴中 Bazhong | 90 | 84 | 93.33 |
| 凉山 Liangshan | 4 | 0 | 0 |
| 资阳 Ziyang | 1 | 0 | 0 |
| 雅安 Ya’an | 56 | 0 | 0 |
| 内江 Neijiang | 12 | 0 | 0 |
| 乐山 Leshan | 18 | 0 | 0 |
| 总数 Total | 490 | 241 | 49.18 |
表3 PDCoV临床样品检测结果
Table 3 Clinical detection of PDCoV samples
| 来源 Origin | 样本数 Sample number | 阳性总数 Positive number | 阳性率 Positive rate/% |
|---|---|---|---|
| 成都 Chengdu | 52 | 34 | 65.38 |
| 南充 Nanchong | 57 | 26 | 45.61 |
| 绵阳 Mianyang | 30 | 12 | 40 |
| 眉山 Meishan | 33 | 3 | 9.09 |
| 遂宁 Suining | 16 | 0 | 0 |
| 德阳 Deyang | 61 | 24 | 39.34 |
| 达州 Dazhou | 27 | 27 | 100 |
| 泸州 Luzhou | 9 | 8 | 88.89 |
| 宜宾 Yibin | 1 | 1 | 100 |
| 阿坝 Aba | 23 | 22 | 95.65 |
| 巴中 Bazhong | 90 | 84 | 93.33 |
| 凉山 Liangshan | 4 | 0 | 0 |
| 资阳 Ziyang | 1 | 0 | 0 |
| 雅安 Ya’an | 56 | 0 | 0 |
| 内江 Neijiang | 12 | 0 | 0 |
| 乐山 Leshan | 18 | 0 | 0 |
| 总数 Total | 490 | 241 | 49.18 |
| [1] | Li G, Chen Q, Harmon KM, et al. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734[J]. Genome Announc, 2014,2(2):e00278-14. |
| [2] |
Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus[J]. J Virol, 2012,86(7):3995-4008.
doi: 10.1128/JVI.06540-11 URL |
| [3] | Marthaler D, Jiang Y, Collins J, et al. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States[J]. Genome Announc, 2014,2(2):e00218-14. |
| [4] |
Wang L, Byrum B, Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014[J]. Emerg Infect Dis, 2014,20(9):1594-1595.
doi: 10.3201/eid2009.140756 URL |
| [5] |
Niederwerder MC, Hesse RA. Swine enteric coronavirus disease:A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada[J]. Transbound Emerg Dis, 2018,65(3):660-675.
doi: 10.1111/tbed.12823 pmid: 29392870 |
| [6] |
Mai K, Feng J, Chen G, et al. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong province in southern China[J]. Transbound Emerg Dis, 2018,65(1):166-173.
doi: 10.1111/tbed.12644 pmid: 28345292 |
| [7] |
Jang G, Lee KK, Kim SH, et al. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016[J]. Transbound Emerg Dis, 2017,64(5):1364-1370.
doi: 10.1111/tbed.12690 pmid: 28758347 |
| [8] |
Saeng-Chuto K, Lorsirigool A, Temeeyasen G, et al. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015[J]. Transbound Emerg Dis, 2017,64(1):3-10.
doi: 10.1111/tbed.12585 pmid: 27718337 |
| [9] |
Suzuki T, Shibahara T, Imai N, et al. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus[J]. Infect Genet Evol, 2018,61:176-182.
doi: 10.1016/j.meegid.2018.03.030 URL |
| [10] |
Dong N, Fang L, Zeng S, et al. Porcine deltacoronavirus in mainland China[J]. Emerg Infect Dis, 2015,21(12):2254-2255.
doi: 10.3201/eid2112.150283 URL |
| [11] |
Wang M, Wang Y, Baloch AR, et al. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China[J]. Transbound Emerg Dis, 2018,65(2):363-369.
doi: 10.1111/tbed.12819 pmid: 29363281 |
| [12] |
Zhang HL, Liang QQ, Li BX, et al. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China[J]. Prev Vet Med, 2019,166:8-15.
doi: 10.1016/j.prevetmed.2019.02.017 URL |
| [13] | Lee S, Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014[J]. Genome Announc, 2014,2(6):e01191-14. |
| [14] | 方谱县, 方六荣, 董楠, 等. 猪δ冠状病毒的研究进展[J]. 病毒学报, 2016,32(2):243-248. |
| Fang PX, Fang LR, Dong N, et al. Research advances in the porcine deltacoronavirus[J]. Chinese Journal of Virology, 2016,32(2):243-248. | |
| [15] | Shang J, Zheng Y, Yang Y, et al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state[J]. J Virol, 2018,92(4):e01556-17. |
| [16] |
Chen R, Fu J, Hu J, et al. Identification of the immunodominant neutralizing regions in the spike glycoprotein of porcine deltacoronavirus[J]. Virus Res, 2020,276:197834.
doi: 10.1016/j.virusres.2019.197834 URL |
| [17] |
Brian DA, Baric RS. Coronavirus genome structure and replication[J]. Curr Top Microbiol Immunol, 2005,287:1-30.
pmid: 15609507 |
| [18] |
Sun DB, Feng L, Shi HY, et al. Spike protein region(aa 636789)of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies[J]. Acta Virol, 2007,51(3):149-156.
pmid: 18076304 |
| [19] |
Carvajal A, Lanza I, Diego R, et al. Evaluation of a blocking ELISA using monoclonal antibodies for the detection of porcine epidemic diarrhea virus and its antibodies[J]. J Vet Diagn Invest, 1995,7(1):60-64.
pmid: 7779966 |
| [20] |
Su M, Li C, Guo D, et al. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus[J]. J Vet Med Sci, 2016,78(4):601-606.
doi: 10.1292/jvms.15-0533 URL |
| [21] | 杨浩, 方六荣, 董楠, 等. 基于猪德尔塔冠状病毒重组核衣壳蛋白的 ELISA抗体检测方法的建立与评价[J]. 微生物学通报, 2017,44(12):2830-2838. |
| Yang H, Fang LR, Dong N, et al. Development and evaluation of an indirect ELISA based on recombinant nucleocapsid protein for detecting antibodies against porcine deltacoronavirus[J]. Microbiology China, 2017,44(12):2830-2838. | |
| [22] |
Luo SX, Fan JH, Opriessnig T, et al. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies[J]. J Virol Methods, 2017,249:76-78.
doi: 10.1016/j.jviromet.2017.08.020 URL |
| [23] | 殷震. 动物病毒学基础[M]. 长春: 吉林人民出版社, 1980. |
| Yin Z. Fundamentals of animal virology[M]. Changchun: Jilin People’s Press, 1980. | |
| [24] | Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014[J]. Emerg Infect Dis, 2014,20(7):1227-1230. |
| [25] | 孙东波. 猪流行性腹泻病毒S蛋白抗原表位鉴定及受体结合域的初步筛选[D]. 北京:中国农业科学院, 2008. |
| Sun DB. Identification of antigenic epitopes and screening receptor binding domain on the S protein of porcine epidemic diarrhea virus[D]. Beijing:Chinese Academy of Agricultural Sciences, 2008. | |
| [26] |
Ma Y, Zhang Y, Liang X, et al. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus[J]. Vet Microbiol, 2016,186:90-96.
doi: 10.1016/j.vetmic.2016.02.004 URL |
| [27] |
Gimenez-Lirola LG, Zhang J, Carrillo-Avila JA, et al. Reactivity of porcine epidemic diarrhea virus structural proteins to antibodies against porcine enteric coronaviruses:diagnostic implications[J]. J Clin Microbiol, 2017,55(5):1426-1436.
doi: 10.1128/JCM.02507-16 URL |
| [28] |
Thachil A, Gerber PF, Xiao CT, et al. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies[J]. PLoS One, 2015,10(4):e0124363.
doi: 10.1371/journal.pone.0124363 URL |
| [29] |
Sestak K, Zhou Z, Shoup DI, et al. Evaluation of the baculovirus-expressed S glycoprotein of transmissible gastroenteritis virus(TGEV)as antigen in a competition ELISA to differentiate porcine respiratory coronavirus from TGEV antibodies in pigs[J]. J Vet Diagn Invest, 1999,11(3):205-214.
pmid: 10353350 |
| [30] |
Gerber PF, Gong Q, Huang YW, et al. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA[J]. Vet J, 2014,202(1):33-36.
doi: 10.1016/j.tvjl.2014.07.018 URL |
| [31] | 王经纬, 雷喜梅, 覃盼, 等. 猪丁型冠状病毒荧光定量 RT-PCR 与 S1 蛋白间接ELISA 检测方法的建立及应用[J]. 生物工程学报, 2017,33(8):1265-1275. |
| Wang JW, Lei XM, Qin P, et al. Development and application of real-time RT-PCR and S1 protein-based indirect ELISA for porcine deltacoronavirus[J]. Chin J Biotech, 2017,33(8):1265-1275. | |
| [32] |
Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins[J]. J Biosci Bioeng, 2005,99(4):303-310.
doi: 10.1263/jbb.99.303 URL |
| [33] | Burgess RR. Refolding solubilized inclusion body proteins[J]. Methods Enzymol, 2009,463:259-282. |
| [34] | 逄凤娇, 俞正玉, 何孔旺, 等. 猪丁型冠状病毒重组N蛋白间接ELISA抗体检测方法的建立[J]. 中国预防兽医学报, 2017,39(6):461-465. |
| Pang FJ, Yu ZY, He KW, et al. Development of an indirect ELISA for detection antibody to porcine deltacoronavirus using recombinant N protein[J]. Chinese Journal of Preventive Veterinary Medicine, 2017,39(6):461-465. |
| [1] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| [2] | 索青青, 吴楠, 杨慧, 李莉, 王锡锋. 水稻咖啡酰辅酶A-O-甲基转移酶基因的原核表达、抗体制备和应用[J]. 生物技术通报, 2022, 38(8): 135-141. |
| [3] | 覃雪晶, 王雨涵, 曹一博, 张凌云. 青杄PwHAP5基因原核表达及多克隆抗体制备[J]. 生物技术通报, 2022, 38(8): 142-149. |
| [4] | 王光丽, 范婵, 王辉, 卢惠芳, 夏灵尹, 黄健, 闵迅. 霍乱弧菌溶血素HlyA的原核表达、纯化及多克隆抗体制备与鉴定[J]. 生物技术通报, 2022, 38(7): 269-277. |
| [5] | 汪巧菊, 胡雨萌, 温亚亚, 宋丽, 孟闯, 潘志明, 焦新安. 新型冠状病毒S1蛋白的表达及活性鉴定[J]. 生物技术通报, 2022, 38(3): 157-163. |
| [6] | 沈俊强, 张莉萍, 于瑞明, 王永录, 潘丽, 刘霞, 刘新生. 猪嵴病毒结构蛋白VP0与VP1原核表达及间接ELISA方法的建立[J]. 生物技术通报, 2022, 38(10): 243-253. |
| [7] | 山草梅, 叶蕾, 张连虎, 况卫刚, 孙晓棠, 马建, 崔汝强. 水稻抗潜根线虫基因OsRAI1的克隆及功能分析[J]. 生物技术通报, 2021, 37(7): 146-155. |
| [8] | 曾福源, 苏泽辉, 周诗慧, 谢妙, 庞欢瑛. 溶藻弧菌PEPCK蛋白原核表达及其乙酰化、琥珀酰化修饰的鉴定[J]. 生物技术通报, 2021, 37(5): 84-91. |
| [9] | 张西西, 张怡青, 李玉林, 韩笑, 王国强, 王晓军, 王旭东, 王云龙. 新型冠状病毒(SARS-CoV-2)N蛋白C端重组蛋白的原核表达、纯化及应用[J]. 生物技术通报, 2021, 37(5): 92-97. |
| [10] | 白福美, 李至敏, 王小琴, 胡紫微, 鲍玲玲, 李志敏. 集胞藻PCC6803中N-乙酰鸟氨酸转氨酶的生化表征及结构分析[J]. 生物技术通报, 2021, 37(5): 98-107. |
| [11] | 彭利忠, 张鹏, 周雯雯, 曾旭辉, 张小宁. 精子特异性蛋白Cabs1多克隆抗体的制备及多用途验证[J]. 生物技术通报, 2021, 37(3): 261-270. |
| [12] | 贺扬, 余巧玲, 王均, 覃川杰, 李华涛. 罗非鱼原核表达基因研究进展[J]. 生物技术通报, 2021, 37(2): 195-202. |
| [13] | 唐禄, 董丽平, 尹茉莉, 刘磊, 董媛, 王会岩. 成纤维细胞生长因子20单克隆抗体的制备及鉴定[J]. 生物技术通报, 2021, 37(10): 179-185. |
| [14] | 段应策, 胡姿仪, 杨帆, 李金涛, 邬向丽, 张瑞颖. 香菇草酰乙酸水解酶基因LeOAH1克隆及表达分析[J]. 生物技术通报, 2020, 36(9): 227-234. |
| [15] | 陈汭, 付嘉钰, 刘浩宇, 李成, 赵玉佳, 胡靖飞, 瞿欢, 曹三杰, 文心田, 文翼平, 赵勤, 伍锐, 黄小波. 猪δ冠状病毒(PDCoV)N蛋白的原核表达及多克隆抗体制备[J]. 生物技术通报, 2020, 36(8): 104-110. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||