








生物技术通报 ›› 2021, Vol. 37 ›› Issue (8): 85-94.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1386
岑由飞1,2( ), 朱牧孜2, 叶伟2, 李赛妮2, 钟国华1(
), 朱牧孜2, 叶伟2, 李赛妮2, 钟国华1( ), 章卫民2(
), 章卫民2( )
)
收稿日期:2020-11-14
出版日期:2021-08-26
发布日期:2021-09-10
作者简介:岑由飞,女,硕士研究生,研究方向:微生物功能基因;E-mail: 基金资助:
CEN You-fei1,2( ), ZHU Mu-zi2, YE Wei2, LI Sai-ni2, ZHONG Guo-hua1(
), ZHU Mu-zi2, YE Wei2, LI Sai-ni2, ZHONG Guo-hua1( ), ZHANG Wei-min2(
), ZHANG Wei-min2( )
)
Received:2020-11-14
Published:2021-08-26
Online:2021-09-10
摘要:
利用染色体步移技术对植物内生真菌Paramyrothecium roridum中单端孢霉烯毒素的生物合成基因tri5、tri12启动子进行克隆,分别利用原核和真核表达系统以及荧光素酶表达系统对启动子核心区域进行鉴定,发现tri12的启动子片段12-f1对氨苄青霉素抗性基因和潮霉素抗性基因表达的启动效率最高,tri5的启动子片段5-f0对荧光素酶基因表达的启动效率最高;同时对启动子的功能组件进行分析,发现tri5含有TATA box和CAAT box,而tri12是TATA-less启动子,但含有CAAT box和GC box。本研究为强启动子的筛选及单端孢霉烯类化合物的高效生物合成奠定基础。
岑由飞, 朱牧孜, 叶伟, 李赛妮, 钟国华, 章卫民. Paramyrothecium roridum中单端孢霉烯毒素生物合成基因启动子的克隆和功能鉴定[J]. 生物技术通报, 2021, 37(8): 85-94.
CEN You-fei, ZHU Mu-zi, YE Wei, LI Sai-ni, ZHONG Guo-hua, ZHANG Wei-min. Cloning and Functional Identification of Trichothecene Mycotoxin Biosynthesis Gene Promoter from Paramyrothecium roridum[J]. Biotechnology Bulletin, 2021, 37(8): 85-94.
| 目的基因Target gene | 反向引物Reverse primer sp1(5'-3') | 反向引物Reverse primer sp2(5'-3') | 反向引物Reverse primer sp3(5'-3') |
|---|---|---|---|
| tri5 | TGTACTTGACGTTCTCCAGCAAC | GGGAGATTCTGAGAGAGATAGATA | TCTGTCTGTCTGATCTGTCTGTC |
| tri12 | GCGTTGGTCAACATGAAGAATGG | ACATCTGCGGCGATGTTTGAGAT | ATCAAACGAGGGCTCCGATAGTA |
表1 目的基因tri5和tri12特异性反向引物序列
Table 1 Specific reverse primer sequences of the target genes tri5 and tri12
| 目的基因Target gene | 反向引物Reverse primer sp1(5'-3') | 反向引物Reverse primer sp2(5'-3') | 反向引物Reverse primer sp3(5'-3') |
|---|---|---|---|
| tri5 | TGTACTTGACGTTCTCCAGCAAC | GGGAGATTCTGAGAGAGATAGATA | TCTGTCTGTCTGATCTGTCTGTC |
| tri12 | GCGTTGGTCAACATGAAGAATGG | ACATCTGCGGCGATGTTTGAGAT | ATCAAACGAGGGCTCCGATAGTA |
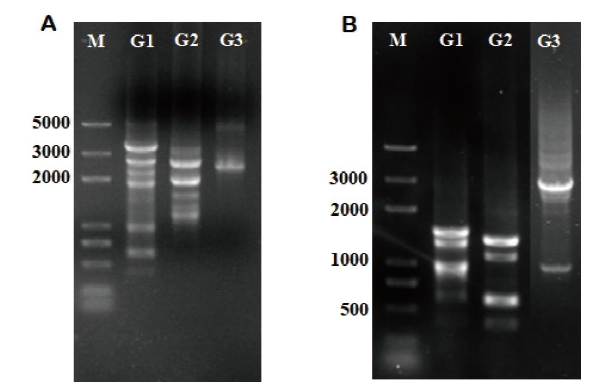
图1 tri5、tri12染色体步移扩增得到的目的片段 M:Marker trans2K plus;A:tri5染色体步移三轮PCR扩增产物;B:tri12染色体步移三轮PCR扩增产物
Fig.1 Target fragments of tri5 and tri12 obtained by chromosome walking M:Marker trans2K plus;A:The PCR amplification products of tri5 after three rounds of chromosome walking;B:The PCR amplification products of tri12 after three rounds of chromosome walking
| 目的基因启动子 Promoter of target gene | 分值 Score | 核心序列 Core sequence |
|---|---|---|
| tri5 | 0.71 | ACCAGAGACGGGAATCACGCGCATT- TCTGGCTACGATATCCCCCCTAACG |
| tri12 | 0.98 | AGGCCCTCGATAAGAAGCGGCGGCG- GGGCCTCGACCAGGAATCGACTACC |
表2 目的基因tri5、tri12启动子核心序列
Table 2 Core sequences of tri5 and tri12 promoters
| 目的基因启动子 Promoter of target gene | 分值 Score | 核心序列 Core sequence |
|---|---|---|
| tri5 | 0.71 | ACCAGAGACGGGAATCACGCGCATT- TCTGGCTACGATATCCCCCCTAACG |
| tri12 | 0.98 | AGGCCCTCGATAAGAAGCGGCGGCG- GGGCCTCGACCAGGAATCGACTACC |
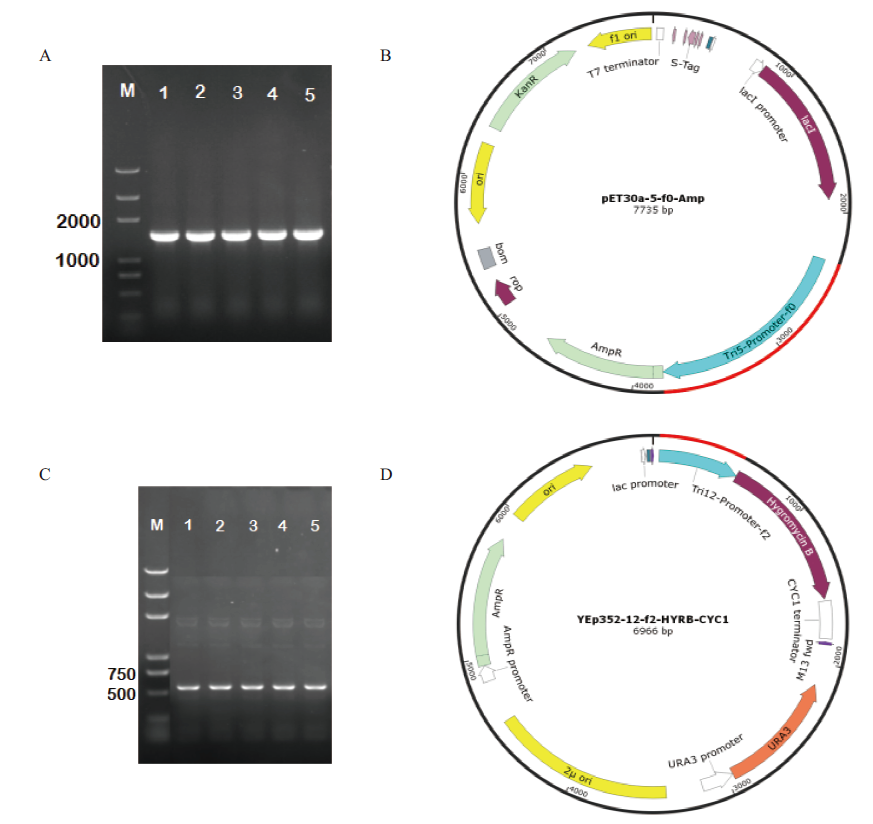
图3 DH5α菌落PCR验证及启动子载体 M:Marker trans2K plus;A:DH5α-pET30a-5-f0-Amp菌落PCR图;B:pET30a-5-f0-Amp载体图谱;C:DH5α-YEp352-12-f2-HYRB-CYC1菌落PCR图;D:YEp352-12-f2-HYRB-CYC1载体图谱
Fig.3 Colony PCR verification of DH5α and promoter vector maps M:Marker trans2K plus. A:Colony PCR of DH5α-pET30a-5-f0-Amp. B:Vector map of pET30a-5-f0-Amp. C:Colony PCR of DH5α-YEp352-12-f2-HYRB-CYC1. D:Vector map of YEp352-12-f2-HYRB-CYC1

图4 不同浓度的氨苄青霉素抗性平板筛选含不同启动子片段的大肠杆菌 A:DH5α-pET30a-5-f0/5-f1/5-f2/12-f0/12-f1/12-f2-Amp点板结果;B:DH5α-pET30a-5-f0/5-f1/5-f2-Amp点板结果;C:DH5α-YEp352-12-f1-HYRB-CYC1和DH5α-pET30a-12-f1-Amp的OD值柱形图。N:DH5α-pET30a-ACP空载(阴性对照);P:DH5α-YEp352-12-f1-HYRB-CYC1(阳性对照);5(f0-f2):DH5α-pET30a-5-f0/5-f1/ 5-f2-Amp;12(f0-f2):DH5α-pET30a-12-f0/12-f1/12-f2-Amp
Fig.4 Screening of E. coli containing different promoter fragments on resistance plates with different concentrations of ampicillin A:The result of culture on plates of DH5α-pET30a-5-f0/5-f1/5-f2/12-f0/12-f1/12-f2-Amp. B:The result of culture on plates of DH5α-pET30a-5-f0/5-f1/5-f2-Amp. C:OD value bar graph of DH5α-YEp352-12-f1-HYRB-CYC1 and DH5α-pET30a-12-f1-Amp. N:DH5α-pET30a-ACP empty carrier(Negative control);P:DH5α-YEp352-12-f1-HYRB-CYC1(Positive control);5(f0-f2):DH5α-pET30a-5-f0/5-f1/ 5-f2-Amp;12(f0-f2):DH5α-pET30a-12-f0/12-f1/12-f2-Amp
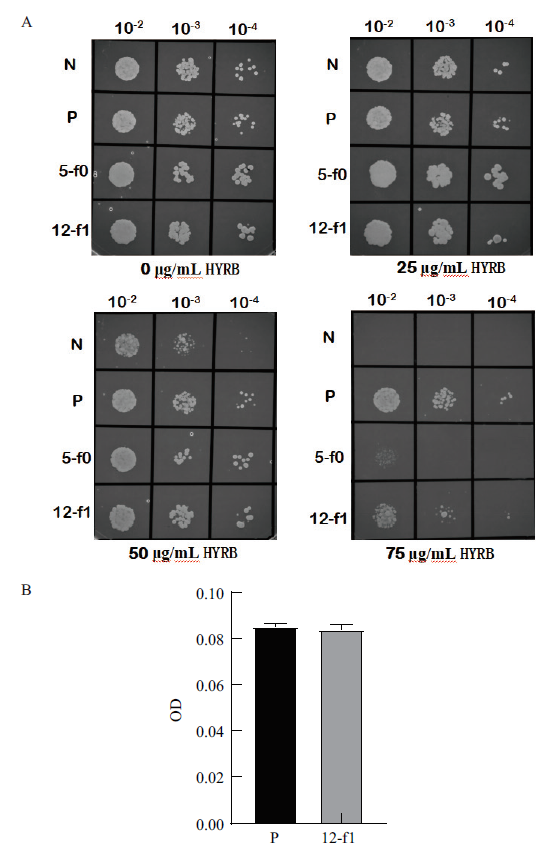
图5 不同浓度的潮霉素抗性平板筛选含不同启动子片段的酿酒酵母 A:BJ5464-YEp352-TEF1-HYRB-CYC1(阳性对照)、BJ5464-YEp352-No pro-HYRB-CYC1(阴性对照)、BJ5464-YEp352-12-f1-HYRB-CYC1和BJ5464-YEp352-5-f0-HYRB-CYC1的点板结果;B:BJ5464-YEp352-TEF1-HYRB-CYC1(阳性对照)和BJ5464-YEp352-12-f1-HYRB-CYC1的OD值柱形图。N:BJ5464-YEp352-No pro-HYRB-CYC1;P:BJ5464-YEp352-TEF1-HYRB-CYC1;5-f0:BJ5464-YEp352-5-f0-HYRB-CYC1;12-f1:BJ5464-YEp352-12-f1-HYRB-CYC1
Fig.5 Screening of S. cerevisiae containing different prom-oter fragments on resistant plates with different concentrations of hygromycin A:The result of culture on plates of BJ5464-YEp352-TEF1-HYRB-CYC1(Positive control),BJ5464-YEp352-No pro-HYRB-CYC1(Negative control),BJ5464-YEp352-12-f1-HYRB-CYC1 and BJ5464-YEp352-5-f0-HYRB-CYC1;B:OD value bar graph of BJ5464-YEp352-TEF1-HYRB-CYC1(Positive control)and BJ5464-YEp352-12-f1-HYRB-CYC1. N:BJ5464-YEp352-No pro-HYRB-CYC1;P:BJ5464-YEp352-TEF1-HYRB-CYC1;5-f0:BJ5464-YEp352-5-f0-HYRB-CYC1;12-f1:BJ5464-YEp352-12-f1-HYRB-CYC1
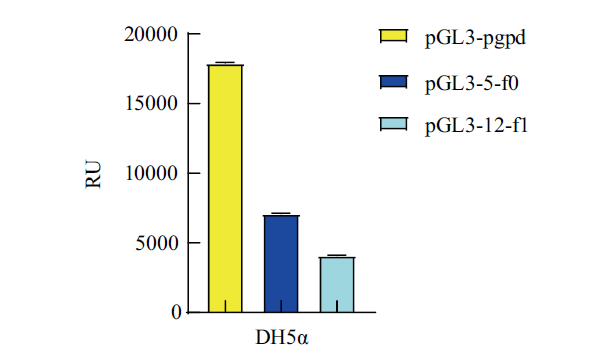
图6 pGL3-5-f0-basic、pGL3-12-f1-basic和pGL3-pgpd-basic载体在DH5α中的荧光素酶活性检测
Fig.6 Luciferase activity detection of pGL3-5-f0-basic,pGL3-12-f1-basic and pGL3-pgpd-basic vectors in DH5α
| [1] | Brown DW, Villani A, Susca A, et al. Gain and loss of a transcription factor that regulates late trichothecene biosynthetic pathway genes in Fusarium[J]. Fungal Genetics and Biology, 2020, 136:553-560. |
| [2] |
Cardoza RE, McCormick SP, Lindo L, et al. A cytochrome P450 monooxygenase gene required for biosynjournal of the trichothecene toxin harzianum A in Trichoderma[J]. Applied Microbiology and Biotechnology, 2019, 103(19):8087-8103.
doi: 10.1007/s00253-019-10047-2 pmid: 31384992 |
| [3] |
Lee SR, Seok S, Ryoo R, et al. Macrocyclic trichothecene mycotoxins from a deadly poisonous mushroom, Podostroma cornu-damae[J]. Journal of Natural Products, 2019, 82(1):122-128.
doi: 10.1021/acs.jnatprod.8b00823 URL |
| [4] |
Nguyen LTT, Jang JY, Kim TY, et al. Nematicidal activity of verrucarin A and roridin A isolated from Myrothecium verrucaria against Meloidogyne incognita[J]. Pesticide Biochemistry and Physiology, 2018, 148:133-143.
doi: S0048-3575(17)30508-4 pmid: 29891364 |
| [5] |
Liu HX, Liu WZ, Chen YC, et al. Cytotoxic trichothecene macrolides from the endophyte fungus Myrothecium roridum[J]. Journal of Asian Natural Products Research, 2016, 18(7):684-692.
doi: 10.1080/10286020.2015.1134505 URL |
| [6] | Ye W, Chen YC, Li HH, et al. Two trichothecene mycotoxins from Myrothecium roridum induce apoptosis of HepG-2 cells via caspase activation and disruption of mitochondrial membrane potential[J]. Molecules, 2016, 21(6):E781. |
| [7] |
Cane DE. Enzymic formation of sesquiterpenes[J]. Chemical Reviews, 1990, 90(7):1089-1103.
doi: 10.1021/cr00105a002 URL |
| [8] |
McCormick SP, Alexander NJ, Trapp SE, et al. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides[J]. Applied and Environmental Microbiology, 1999, 65(12):5252-5256.
pmid: 10583973 |
| [9] |
Proctor RH, McCormick SP, Kim HS, et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi[J]. PLoS Pathogens, 2018, 14(4):e1006946.
doi: 10.1371/journal.ppat.1006946 URL |
| [10] |
Trassaert M, Vandermies M, Carly F, et al. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica[J]. Microbial Cell Factories, 2017, 16(1):141.
doi: 10.1186/s12934-017-0755-0 pmid: 28810867 |
| [11] |
Cooper SJ, Trinklein ND, Anton ED, et al. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome[J]. Genome Research, 2006, 16(1):1-10.
pmid: 16344566 |
| [12] |
Even DY, Kedmi A, Ideses D, et al. Functional screening of core promoter activity[J]. Methods in Molecular Biology, 2017, 1651:77-91.
doi: 10.1007/978-1-4939-7223-4_7 pmid: 28801901 |
| [13] |
Balabanova LA, Shkryl YN, Slepchenko LV, et al. Development of host strains and vector system for an efficient genetic transformation of filamentous fungi[J]. Plasmid, 2019, 101:1-9.
doi: S0147-619X(18)30089-1 pmid: 30465791 |
| [14] | 朱利, 王义, 殷金瑶, 等. 橡胶树白粉菌内源强启动子WY193的克隆及功能鉴定[J]. 分子植物育种, 2020, 18(9):2865-2871. |
| Zhu L, Wang Y, Yin JY, et al. Cloning and function identification of endogenous strong promoter WY193 of Oidium heveae steinm[J]. Molecular Plant Breeding, 2020, 18(9):2865-2871. | |
| [15] | 阎隆飞, 张玉麟. 分子生物学[M]. 第2 版. 北京: 中国农业大学出版社, 1997. |
| Yan LF, Zhang YL. Molecular biology[M]. 2rd ed. Beijing: China Agricultural University Press, 1997. | |
| [16] | Ye W, Liu TM, Zhu MZ, et al. De Novo transcriptome analysis of plant pathogenic fungus Myrothecium roridum and identification of genes associated with trichothecene mycotoxin biosynjournal[J]. International Journal of Molecular Sciences, 2017, 18(3):E497. |
| [17] | Ashrafi M, Fardsi M, Mirshamsi A, et al. Isolation and sequence analysis of gpdII promoter of the white button mushroom(Agaricus bisporus)from strains Holland737 and IM008[J]. International Journal of Horticultural Science and Technology, 2015, 2(1):33-41. |
| [18] |
Madhavan A, Pandey A, et al. Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis[J]. Bioresource Technology, 2017, 245:1334-1342.
doi: S0960-8524(17)30808-8 pmid: 28578805 |
| [19] |
Keränen S, Penttilä M. Production of recombinant proteins in the filamentous fungus Trichoderma reesei[J]. Current Opinion in Biotechnology, 1995, 6(5):534-537.
pmid: 7579664 |
| [20] | 黄自磊, 章卫民, 叶伟, 等. 深海真菌Dichotomomyces cejpii胶霉毒素生物合成基因启动子的克隆和功能鉴定[J]. 生物技术通报, 2018, 34(4):144-150. |
| Huang ZL, Zhang WM, Ye W, et al. Cloning and identification of gene promoter for gliotoxin biosynjournal from deep-sea-derived fungus Dichotomomyces cejpii[J]. Biotechnology Bulletin, 2018, 34(4):144-150. |
| [1] | 周璐祺, 崔婷茹, 郝楠, 赵雨薇, 赵斌, 刘颖超. 化学蛋白质组学在天然产物分子靶标鉴定中的应用[J]. 生物技术通报, 2023, 39(9): 12-26. |
| [2] | 刘玉玲, 王梦瑶, 孙琦, 马利花, 朱新霞. 启动子RD29A对转雪莲SikCDPK1基因烟草抗逆性的影响[J]. 生物技术通报, 2023, 39(9): 168-175. |
| [3] | 江海溶, 崔若琪, 王玥, 白淼, 张明露, 任连海. NH3和H2S降解功能菌的分离鉴定及降解特性研究[J]. 生物技术通报, 2023, 39(9): 246-254. |
| [4] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [5] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [6] | 饶紫环, 谢志雄. 一株Olivibacter jilunii 纤维素降解菌株的分离鉴定与降解能力分析[J]. 生物技术通报, 2023, 39(8): 283-290. |
| [7] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [8] | 马俊秀, 吴皓琼, 姜威, 闫更轩, 胡基华, 张淑梅. 蔬菜软腐病菌广谱拮抗细菌菌株筛选鉴定及防效研究[J]. 生物技术通报, 2023, 39(7): 228-240. |
| [9] | 谢东, 汪流伟, 李宁健, 李泽霖, 徐子航, 张庆华. 一株多功能菌株的发掘、鉴定及解磷条件优化[J]. 生物技术通报, 2023, 39(7): 241-253. |
| [10] | 游子娟, 陈汉林, 邓辅财. 鱼皮生物活性肽的提取及功能活性研究进展[J]. 生物技术通报, 2023, 39(7): 91-104. |
| [11] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [12] | 王一帆, 候林慧, 常永春, 杨亚杰, 陈天, 赵祝跃, 荣二花, 吴玉香. 陆地棉与拟似棉异源六倍体的合成与性状鉴定[J]. 生物技术通报, 2023, 39(5): 168-176. |
| [13] | 车永梅, 郭艳苹, 刘广超, 叶青, 李雅华, 赵方贵, 刘新. 菌株C8和B4的分离鉴定及其耐盐促生效果和机制[J]. 生物技术通报, 2023, 39(5): 276-285. |
| [14] | 陈晓萌, 张雪静, 张欢, 张宝江, 苏艳. 重组牛乳源金黄色葡萄球菌GapC蛋白优势B细胞抗原表位的预测和筛选[J]. 生物技术通报, 2023, 39(5): 306-313. |
| [15] | 赖瑞联, 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺. 猕猴桃过氧化氢酶基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2023, 39(4): 136-147. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||