








生物技术通报 ›› 2021, Vol. 37 ›› Issue (10): 72-80.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0124
卫晓博1,3( ), 侯颖1,2(
), 侯颖1,2( ), 程豪杰1,2, 秦翠丽1,3, 牛明福1,2, 徐建强4(
), 程豪杰1,2, 秦翠丽1,3, 牛明福1,2, 徐建强4( )
)
收稿日期:2021-01-29
出版日期:2021-10-26
发布日期:2021-11-12
作者简介:卫晓博,男,硕士研究生,研究方向:环境微生物学;E-mail: 基金资助:
WEI Xiao-bo1,3( ), HOU Ying1,2(
), HOU Ying1,2( ), CHENG Hao-jie1,2, QIN Cui-li1,3, NIU Ming-fu1,2, XU Jian-qiang4(
), CHENG Hao-jie1,2, QIN Cui-li1,3, NIU Ming-fu1,2, XU Jian-qiang4( )
)
Received:2021-01-29
Published:2021-10-26
Online:2021-11-12
摘要:
苯酚是最常见的有机污染物,为了获得降解苯酚的细菌,并探究其降解苯酚的特性和途径,采用连续富集和稀释平板分离,从农药厂活性污泥中得到一株能以苯酚作为唯一碳源生长的菌株BF-6,通过 16S rRNA 序列分析和生理生化试验将该菌株鉴定为假黄单胞菌(Pseudoxanthomonas sp.)。单因素试验结果表明,菌株BF-6在温度为20-37℃和pH5.0-10.0范围内均可很好的降解苯酚;在此条件下,5%接种量的菌株BF-6在36 h内可将100 mg/L苯酚降解97.79%,96 h可将200 mg/L苯酚降解97.58%。经苯酚诱导后,菌株BF-6细胞粗酶液表现出苯酚羟化酶和邻苯二酚1,2-双加氧酶活性,表明菌株BF-6降解苯酚的途径为邻位途径。本研究为低浓度苯酚的微生物降解提供了新的菌种资源。
卫晓博, 侯颖, 程豪杰, 秦翠丽, 牛明福, 徐建强. 一种苯酚降解菌Pseudoxanthomonas sp. BF-6的分离鉴定及其降解特性及途径研究[J]. 生物技术通报, 2021, 37(10): 72-80.
WEI Xiao-bo, HOU Ying, CHENG Hao-jie, QIN Cui-li, NIU Ming-fu, XU Jian-qiang. Isolation,Identification of Phenol-degrading Pseudoxanthomonas sp. BF-6 and Its Degradation Characteristics and Pathway[J]. Biotechnology Bulletin, 2021, 37(10): 72-80.
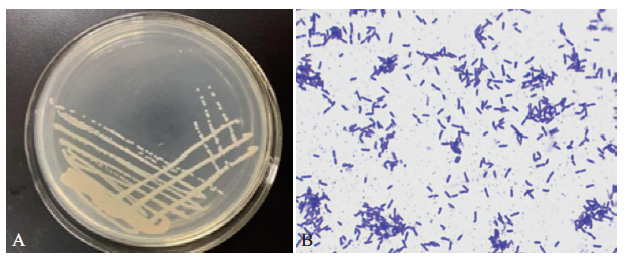
图1 菌株BF-6的菌落和菌体形态 A:菌株BF-6在LB平板上形成的菌落;B:菌株BF-6光学显微镜下形态(1 000×)
Fig.1 Colony and morphology of strain BF-6 A:Colonies of strain BF-6 on LB plate;B:morphology of strain BF-6 under optical microscope(1000×)
| 项目Item | 结果Result | 项目Item | 结果Result |
|---|---|---|---|
| 葡萄糖Glucose | + | 头孢克肟Cefixime | + |
| 蔗糖Sucrose | + | 头孢克洛Cefaclor | + |
| 乳糖Lactose | + | 头孢拉定Cefradine | + |
| 木糖Xylose | - | 呋喃唑酮Furazolidone | + |
| 甘露醇Mannitol | - | 杆菌肽Bacitracin | + |
| 甲醇Methanol | - | 阿奇霉素Azithromycin | - |
| 乙醇Ethanol | + | 四环素Tetracycline | - |
| 牛肉膏Beef paste | + | 卡那霉素Kanamycin | - |
| 蛋白胨Peptone | + | 诺氟沙星Norfloxacin | - |
| 酵母粉Yeast powder | + | 革兰氏染色Gram stain | - |
| 尿素Urea | + | 甲基红试验Methyl red test | - |
| (NH4)2SO4 | + | V-P试验V-P test | - |
| KNO3 | + | 过氧化氢酶试验Catalase test | + |
| NaNO2 | + | 淀粉水解酶试验Starch hydrolase test | - |
| 青霉素Penicillin | + | 反硝化试验Denitrification experiment | - |
| 阿莫西林Amoxicillin | + | 明胶液化试验Gelatin liquefaction test | - |
| 氨苄西林Ampicillin | + | 最适生长温度Optimal growth temperature | 37℃ |
| 链霉素Streptomycin | + | 最适生长pH Optimal growth pH | 8.0 |
表1 菌株BF-6部分生理生化试验结果
Table 1 Results of physiological and biochemical test of strain BF-6
| 项目Item | 结果Result | 项目Item | 结果Result |
|---|---|---|---|
| 葡萄糖Glucose | + | 头孢克肟Cefixime | + |
| 蔗糖Sucrose | + | 头孢克洛Cefaclor | + |
| 乳糖Lactose | + | 头孢拉定Cefradine | + |
| 木糖Xylose | - | 呋喃唑酮Furazolidone | + |
| 甘露醇Mannitol | - | 杆菌肽Bacitracin | + |
| 甲醇Methanol | - | 阿奇霉素Azithromycin | - |
| 乙醇Ethanol | + | 四环素Tetracycline | - |
| 牛肉膏Beef paste | + | 卡那霉素Kanamycin | - |
| 蛋白胨Peptone | + | 诺氟沙星Norfloxacin | - |
| 酵母粉Yeast powder | + | 革兰氏染色Gram stain | - |
| 尿素Urea | + | 甲基红试验Methyl red test | - |
| (NH4)2SO4 | + | V-P试验V-P test | - |
| KNO3 | + | 过氧化氢酶试验Catalase test | + |
| NaNO2 | + | 淀粉水解酶试验Starch hydrolase test | - |
| 青霉素Penicillin | + | 反硝化试验Denitrification experiment | - |
| 阿莫西林Amoxicillin | + | 明胶液化试验Gelatin liquefaction test | - |
| 氨苄西林Ampicillin | + | 最适生长温度Optimal growth temperature | 37℃ |
| 链霉素Streptomycin | + | 最适生长pH Optimal growth pH | 8.0 |
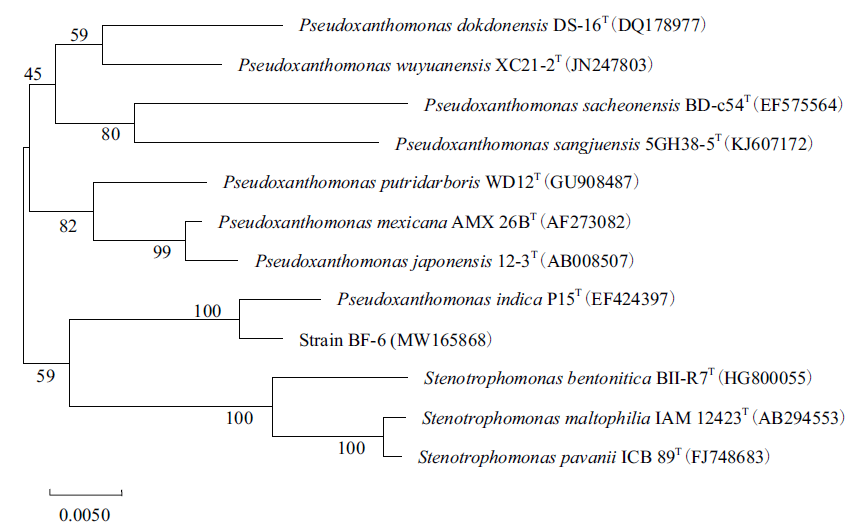
图2 基于16S rRNA基因序列构建的菌株BF-6系统发育树
Fig.2 Phylogenetic tree constructed by the neighbor-joining analysis based on 16S rRNA gene sequences of strain BF-6 and related species
| [1] | Jiang Y, Shang Y, Yang K, et al. Phenol degradation by halophilic fungal isolate JS4 and evaluation of its tolerance of heavy metals[J]. Appl MicrobiolBiot, 2016, 100(4):1883-1890. |
| [2] | 苏琼, 江子骏. 高效苯酚降解菌的筛选及其降解特性分析[J]. 湖北大学学报:自然科学版, 2019, 41(6):567-571. |
| Su Q, Jiang ZJ. Screening and degrading property of highly effective phenol degrading bacterium strain[J]. J Hubei Univ:Nat Sci, 2019, 41(6):567-571. | |
| [3] | 贺强礼, 刘文斌, 杨海君, 等. 一株苯酚降解菌的筛选鉴定及响应面法优化其降解[J]. 环境科学学报, 2016, 36(1):112-123. |
|
He QL, Liu WB, Yang HJ, et al. Isolation, identification of a phenol-degradaing strain and optimization for phenol degradation using response surface methodology[J]. J Environ Sci-China, 2016, 36(1):112-123.
doi: 10.1016/j.jes.2015.03.032 URL |
|
| [4] | 陈焕, 方雅瑜, 尹晓辉, 等. 苯酚降解菌的研究进展[J]. 安徽农业科学, 2015, 43(5):201-205. |
| Chen H, Fang YY, Yin XH, et al. Research progress of phenol degradation bacteria[J]. J Anhui Agricul Sci, 2015, 43(5):201-205. | |
| [5] | Mohanty SS, Jena HM. Biodegradation of phenol by free and immobilized cells of a novel Pseudomonas sp NBM11[J]. Braz JChem Eng, 2017, 34(1):75-84. |
| [6] | Singh U, Arora NK, Sachan P. Simultaneous biodegradation of phenol and cyanide present in coke-oven effluent using immobilized Pseudomonas putida and Pseudomonas stutzeri[J]. Braz JMicrobiol, 2018, 49(1):38-44. |
| [7] | Hamdi H, Hellal A. Optimization of phenol biodegradation by immobilized Bacillus subtilis isolated from hydrocarbons-contaminated water using the factorial design methodology[J]. JSerb Chem Soc, 2019, 84(7):679-688. |
| [8] |
Ke Q, Zhang YG, Wu XL, et al. Sustainable biodegradation of phenol by immobilized Bacillus sp SAS19 with porous carbonaceous gels as carriers[J]. J Environ Manage, 2018, 222:185-189.
doi: 10.1016/j.jenvman.2018.05.061 URL |
| [9] | Van Dexter S, Boopathy R. Biodegradation of phenol by Acinetobacter tandoii isolated from the gut of the termite[J]. EnvironSci Pollut R, 2018, 26(33):34067-34072. |
| [10] | 张海涛, 刘文斌, 杨海君, 等. 一株耐盐高效苯酚降解菌的筛选、鉴定、响应面法优化与降酚动力学研究[J]. 环境科学学报, 2016, 36(9):3200-3207. |
| Zhang HT, Liu WB, Yang HJ, et al. Isolation, identification, optimization via response surface methodology of a highly-efficient phenol-degrading salt-tolerant strain and its degradation kinetics[J]. J Environ Sci-China, 2016, 36(9):3200-3207. | |
| [11] |
Zhao TT, Gao YH, Yu TT, et al. Biodegradation of phenol by a highly tolerant strain Rhodococcus ruber C1:biochemical characterization and comparative genome analysis[J]. Ecotox Environ Safe, 2021, 208:111709.
doi: 10.1016/j.ecoenv.2020.111709 URL |
| [12] | 马馨月, 魏思洁, 柯檀, 等. 红球菌PB-1对高浓度苯酚及苯系物的降解研究[J]. 安全与环境工程, 2020, 27(2):85-91. |
| Ma XY, Wei SJ, Ke T, et al. Degradation of high concentration phenol and benzenes by Rhodococcus sp. PB-1[J]. Safe Environ Eng, 2020, 27(2):85-91. | |
| [13] |
Azadi D, Shojaei H. Biodegradation of polycyclic aromatic hydrocarbons, phenol and sodium sulfate by Nocardia species isolated and characterized from Iranian ecosystems[J]. Sci Rep, 2020, 10:21860.
doi: 10.1038/s41598-020-78821-1 URL |
| [14] |
Silva NCGE, de Macedon AC, Pinheiro ADT, et al. Phenol biodegradation by Candida tropicalis ATCC 750 immobilized on cashew apple bagasse[J]. J Environ Chem Eng, 2019, 7(3):103076.
doi: 10.1016/j.jece.2019.103076 URL |
| [15] |
Patel A, Sartaj K, Arora N, et al. Biodegradation of phenol via meta cleavage pathway triggers de novo TAG biosynjournal pathway in oleaginous yeast[J]. J Hazard Mater, 2017, 340:47-56.
doi: S0304-3894(17)30512-5 pmid: 28711832 |
| [16] |
Gerginova M, Manasiev J, Yemendzhiev H, et al. Biodegradation of phenol by antarctic strains of Aspergillus fumigatus[J]. Z Naturforsch C, 2013, 68(9-10):384-393.
pmid: 24459772 |
| [17] |
Filipowicz N, Momotko M, Boczkaj G, et al. Determination of phenol biodegradation pathways in three psychrotolerant yeasts, Candida subhashii A01(1), Candida oregonensis B02(1)and Schizoblastosporion starkeyi-henricii L01(2), isolated from Rucianka peatland[J]. Enzyme Microb Tech, 2020, 141:109663.
doi: 10.1016/j.enzmictec.2020.109663 pmid: 33051016 |
| [18] | 东秀珠, 蔡妙英. 常见细菌系统鉴定手册[M]. 北京: 科学出版社, 2001:370-385. |
| Dong XZ, Cai MY. Identification methods of common bacteria[M]. Beijing: Science Press, 2001:370-385. | |
| [19] | 国家环保局. 水质挥发酚的测定4-氨基安替比林分光光度法(HJ 503-2009)[S]. 北京: 中国环境科学出版社, 2009:5-7. |
| National Environmental Protection Agency, Water quality-Determination of volatile phenol 4-aminoantipyrine spectrophotometric method(HJ 503-2009)[S], Beijing: China Environmental Science Press, 2009:5-7. | |
| [20] |
Setlhare B, Kumar A, Mokoena MP, et al. Phenol hydroxylase from Pseudomonas sp. KZNSA:Purification, characterization and prediction of three-dimensional structure[J]. Int J Biol Macromol, 2020, 146:1000-1008.
doi: 10.1016/j.ijbiomac.2019.09.224 URL |
| [21] |
Wu LY, Ali DC, Liu P, et al. Degradation of phenol via ortho-pathway by Kocuria sp. strain TIBETAN4 isolated from the soils around Qinghai Lake in China[J]. PLoS One, 2018, 13(6):e0199572.
doi: 10.1371/journal.pone.0199572 URL |
| [22] | Kotresha D, Vidyasagar GM. Chromosomally encoded phenol hydroxylase gene for degradation of phenol by Pseudomonas aeruginosa MTCC 4996[J]. Int J Micro Res Tech, 2014, 2(2):1-7. |
| [23] | 赵化冰, 覃琨, 谭之磊, 等. 邻苯二酚1, 2-双加氧酶的结构、功能及同工酶现象研究进展[J]. 微生物学通报, 2019, 46(9):2419-2425. |
| Zhao HB, Qin K, Tan ZL, et al. Advances in study on structure, enzymatic property and isoenzymes of catechol 1, 2-dioxygenases[J]. Microbiol China, 2019, 46(9):2419-2425. |
| [1] | 饶紫环, 谢志雄. 一株Olivibacter jilunii 纤维素降解菌株的分离鉴定与降解能力分析[J]. 生物技术通报, 2023, 39(8): 283-290. |
| [2] | 游子娟, 陈汉林, 邓辅财. 鱼皮生物活性肽的提取及功能活性研究进展[J]. 生物技术通报, 2023, 39(7): 91-104. |
| [3] | 车永梅, 郭艳苹, 刘广超, 叶青, 李雅华, 赵方贵, 刘新. 菌株C8和B4的分离鉴定及其耐盐促生效果和机制[J]. 生物技术通报, 2023, 39(5): 276-285. |
| [4] | 王凤婷, 王岩, 孙颖, 崔文婧, 乔凯彬, 潘洪玉, 刘金亮. 耐盐碱土曲霉SYAT-1的分离鉴定及抑制植物病原真菌特性研究[J]. 生物技术通报, 2023, 39(2): 203-210. |
| [5] | 张傲洁, 李青云, 宋文红, 颜少慧, 唐爱星, 刘幽燕. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303. |
| [6] | 李柳, 穆迎春, 刘璐, 张洪玉, 徐锦华, 杨臻, 乔璐, 宋金龙. 氟喹诺酮类抗生素及耐药基因污染控制的研究进展[J]. 生物技术通报, 2022, 38(9): 84-95. |
| [7] | 王亚军, 司运美. 除磷菌CP-7的筛选及其降解特性研究[J]. 生物技术通报, 2022, 38(7): 258-268. |
| [8] | 祖雪, 周瑚, 朱华珺, 任佐华, 刘二明. 枯草芽孢杆菌K-268的分离鉴定及对水稻稻瘟病的防病效果[J]. 生物技术通报, 2022, 38(6): 136-146. |
| [9] | 王春艳, 腊贵晓, 苏秀红, 李萌, 董诚明. 地黄不同时期内生促生细菌的筛选及其促生特性分析[J]. 生物技术通报, 2022, 38(4): 242-252. |
| [10] | 张功友, 王一涵, 郭敏, 张婷婷, 王兵, 刘红美. 重楼中一株产纤维素酶内生真菌的分离及鉴定[J]. 生物技术通报, 2022, 38(2): 95-104. |
| [11] | 牛鸿宇, 舒倩, 杨海君, 颜智勇, 谭菊. 一株十二烷基硫酸钠高效降解菌的分离鉴定、降解特性及代谢途径研究[J]. 生物技术通报, 2022, 38(12): 287-299. |
| [12] | 崔欣雨, 李荣荣, 蔡瑞, 王妍, 郑猛虎, 徐春城. 苜蓿青贮中乳酸降解菌的分离、鉴定及降解性能研究[J]. 生物技术通报, 2021, 37(9): 58-67. |
| [13] | 汪颖, 陈永静, 孙庆业, 杨梦瑶, 吴盾. Inquilinus sp. P6-4菌株的生理生化特征及萘降解特性[J]. 生物技术通报, 2021, 37(6): 117-126. |
| [14] | 沈聪, 刘爽, 王春霞, 严雪梅, 代金霞. 盐池采油区污染土壤石油降解菌的筛选鉴定及其降解特性[J]. 生物技术通报, 2021, 37(6): 127-135. |
| [15] | 李瑾, 彭可为, 潘求一, 朱哲远, 彭迪. 解淀粉芽胞杆菌HR-2的分离鉴定及对水稻稻瘟病菌的拮抗效果[J]. 生物技术通报, 2021, 37(3): 27-34. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||