








生物技术通报 ›› 2022, Vol. 38 ›› Issue (2): 173-183.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0345
寇航1,2( ), 王艳梅1, 李彤2, 薄明井2, 张惟材2, 熊向华2(
), 王艳梅1, 李彤2, 薄明井2, 张惟材2, 熊向华2( ), 黎明1(
), 黎明1( )
)
收稿日期:2021-03-20
出版日期:2022-02-26
发布日期:2022-03-09
作者简介:寇航,男,硕士研究生,研究方向:微生物发酵;E-mail: 基金资助:
KOU Hang1,2( ), WANG Yan-mei1, LI Tong2, BO Ming-jing2, ZHANG Wei-cai2, XIONG Xiang-hua2(
), WANG Yan-mei1, LI Tong2, BO Ming-jing2, ZHANG Wei-cai2, XIONG Xiang-hua2( ), LI Ming1(
), LI Ming1( )
)
Received:2021-03-20
Published:2022-02-26
Online:2022-03-09
摘要:
通过构建Methylovorus sp. J1-1基因组尺度代谢网络模型(genome scale of metabolic network model,GSMM),发掘能够提升吡咯喹啉醌(pyrroloquinoline quinone,PQQ)产量的发酵策略和相关靶基因。基于其注释的基因组和生化信息,构建J1-1的GSMM。再通过COBRApy预测可能使PQQ产量提高的氨基酸和潜在的靶基因并进行验证。构建了Methylovorus sp. J1-1的GSMM模型iKH584,共有584个基因,779个生化反应,121个交换反应与765个代谢产物,且可以用于后续模拟。根据iKH584模拟,外源添加谷氨酸、谷氨酰胺和脯氨酸、过表达glyA与hps1以及敲除hps2基因,均具有促进PQQ合成效果。结果表明:添加谷氨酸与脯氨酸时,PQQ的产量分别提高了10.4%与22.9%;过表达基因glyA与hps1,PQQ产量分别提高20.6%与14.6%;敲除基因hps2,PQQ产量最高可达140.84 mg/L,提高了8.0%,这与模拟结果基本一致,表明构建的模型基本正确。最后,在5 L发酵罐进行J1-1△hps2的分批补料发酵,PQQ的产量为812.64 mg/L,比亲本菌株提高了11.1%。建立的模型可以用来指导J1-1的发酵和菌株改造,提高PQQ产量。
寇航, 王艳梅, 李彤, 薄明井, 张惟材, 熊向华, 黎明. 基于Methylovorus sp. J1-1基因组尺度代谢网络优化吡咯喹啉醌合成[J]. 生物技术通报, 2022, 38(2): 173-183.
KOU Hang, WANG Yan-mei, LI Tong, BO Ming-jing, ZHANG Wei-cai, XIONG Xiang-hua, LI Ming. Fermentation Optimization for PQQ Synthesis Based on the Genome-scale Metabolic Model of Methylovorus sp. J1-1[J]. Biotechnology Bulletin, 2022, 38(2): 173-183.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物功能Primer function |
|---|---|---|
| glyA-F | GCCTGCAGGTCGACTCTAGATATAGCGCTCAACAAGGACCTC | glyA基因扩增 Gene amplification of glyA |
| glyA-R | GTGAATTCGAGCTCGGTACCTTAAGCGCCGTATACCGGG | |
| hps1-F | GCCTGCAGGTCGACTCTAGATATTGCTGACTTGAATAGCGCTAAT | hps1基因扩增 Gene amplification of hps1 |
| hps1-R | GTGAATTCGAGCTCGGTACCTTAGTGAGCCAGCGAAGTGA | |
| hps2-up-F | AGAAAAGATCAAAGGATCTTCGGATCCTGCGGAATTCCTTTTGTGG | hps2基因敲除质粒构建 Construction of hps2 knockout plasmid |
| hps2-up-R | CACTATAGGGCGAATTGCTCGAGTGAAGTGGCACAGAACCAG | |
| hps2-kan-F | CCCACAAAAGGAATTCCGCAGGATCCGAAGATCCTTTGATCTTTTC | |
| hps2-kan-R | TAAAGCTCAGCGGGGCTGATACTAGTCAGGTGGCACTTTTCGG | |
| hps2-down-F | TTCCCCGAAAAGTGCCACCTGACTAGTATCAGCCCCGCTGAGC | |
| hps2-down-R | GAGCTCCACCGCGGTGGCGGCCGCATTGCCTTGCTGGGGG | |
| hps2-CF | GCACCATGGGCCAGTCTGA | hps2敲除菌株鉴定 Identifying the strain with hps2 knockout |
| hps2-CR | GGAGCACTTTCACCAGAAGCT | |
| glyA-Y-F | AGTGGATGCTGGTGCCAACAT | 实时荧光定量PCR qRT-PCR |
| glyA-Y-R | GCCGCTGGAAGTCGAACCTT | |
| hps1-Y-F | ATCGGTTCGCCAGCCATCAC | |
| hps1-Y-R | GCCTTGGTCGCAGCAATCAC | |
| fdh-Y-F | AGCTGCACGTACCAAGGAAGTT | |
| fdh-Y-R | TGTTGGCACCAGCATCCACTAC | |
| fae-Y-F | GCCGCTGGAAGTCGAACCTT | |
| fae-Y-R | GCCACGCAGGTTGATCCACTT |
表1 本实验所用引物
Table 1 Primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物功能Primer function |
|---|---|---|
| glyA-F | GCCTGCAGGTCGACTCTAGATATAGCGCTCAACAAGGACCTC | glyA基因扩增 Gene amplification of glyA |
| glyA-R | GTGAATTCGAGCTCGGTACCTTAAGCGCCGTATACCGGG | |
| hps1-F | GCCTGCAGGTCGACTCTAGATATTGCTGACTTGAATAGCGCTAAT | hps1基因扩增 Gene amplification of hps1 |
| hps1-R | GTGAATTCGAGCTCGGTACCTTAGTGAGCCAGCGAAGTGA | |
| hps2-up-F | AGAAAAGATCAAAGGATCTTCGGATCCTGCGGAATTCCTTTTGTGG | hps2基因敲除质粒构建 Construction of hps2 knockout plasmid |
| hps2-up-R | CACTATAGGGCGAATTGCTCGAGTGAAGTGGCACAGAACCAG | |
| hps2-kan-F | CCCACAAAAGGAATTCCGCAGGATCCGAAGATCCTTTGATCTTTTC | |
| hps2-kan-R | TAAAGCTCAGCGGGGCTGATACTAGTCAGGTGGCACTTTTCGG | |
| hps2-down-F | TTCCCCGAAAAGTGCCACCTGACTAGTATCAGCCCCGCTGAGC | |
| hps2-down-R | GAGCTCCACCGCGGTGGCGGCCGCATTGCCTTGCTGGGGG | |
| hps2-CF | GCACCATGGGCCAGTCTGA | hps2敲除菌株鉴定 Identifying the strain with hps2 knockout |
| hps2-CR | GGAGCACTTTCACCAGAAGCT | |
| glyA-Y-F | AGTGGATGCTGGTGCCAACAT | 实时荧光定量PCR qRT-PCR |
| glyA-Y-R | GCCGCTGGAAGTCGAACCTT | |
| hps1-Y-F | ATCGGTTCGCCAGCCATCAC | |
| hps1-Y-R | GCCTTGGTCGCAGCAATCAC | |
| fdh-Y-F | AGCTGCACGTACCAAGGAAGTT | |
| fdh-Y-R | TGTTGGCACCAGCATCCACTAC | |
| fae-Y-F | GCCGCTGGAAGTCGAACCTT | |
| fae-Y-R | GCCACGCAGGTTGATCCACTT |
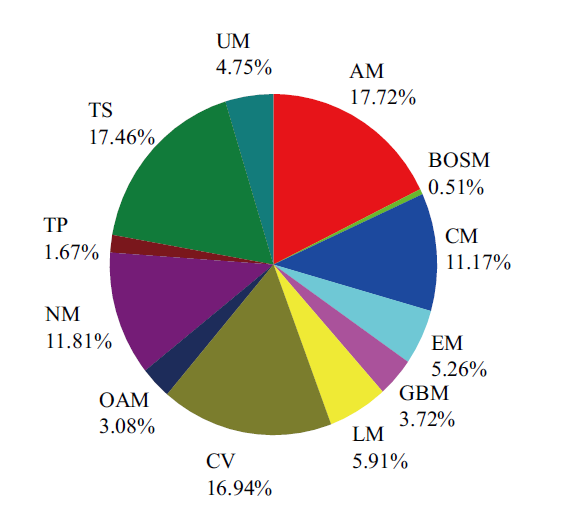
图1 基因组代谢网络的反应分布 AM(氨基酸代谢);BOSM(其他次级代谢);CM(碳水化合物代谢);EM(能量代谢);GBM(糖合成代谢);LM(脂质代谢);CV(辅因子和维生素代谢);OAM(其他氨基酸代谢);NM(核苷酸代谢);TP(萜类和多酮类代谢);TS(运输系统);UM(未分类反应)
Fig. 1 Distributions of reactions in GSMM AM(Amino Acids Metabolism),BOSM(Biosynthesis of Other Secondary Metabolism),CM(Carbohydrates Metabolism),EM(Energy Metabolism),GBM(Glycan Synthesis and Metabolism),LM(Lipids Metabolism),CV(Cofactors and Vitamins Metabolism),OAM(Other Amino Acid Metabolism),NM(Nucl-eotides Metabolism),TP(Terpenoids and Polyketides),TS(Transport system),and UM(Unclassified Reactions)
| 底物Substrate | 实验值In vivo | 模拟值In silico |
|---|---|---|
| 碳源 Carbon source | ||
| 葡萄糖 Glucose | + | + |
| 甲醇 Methanol | + | + |
| 乙醇 Ethanol | + | + |
| 果糖 Fructose | + | + |
| 氮源 Nitrogen source | ||
| 尿素 Urea | + | + |
| 硝酸钠 Sodium nitrate | + | + |
| 硝酸铵 Ammonium nitrate | + | + |
| 氯化铵 Ammonium chloride | + | + |
| 硫酸铵 Ammonium sulfate | + | + |
表2 J1-1在不同碳源和氮源上的生长模拟
Table 2 Simulation of growing in carbon and nitrogen sources
| 底物Substrate | 实验值In vivo | 模拟值In silico |
|---|---|---|
| 碳源 Carbon source | ||
| 葡萄糖 Glucose | + | + |
| 甲醇 Methanol | + | + |
| 乙醇 Ethanol | + | + |
| 果糖 Fructose | + | + |
| 氮源 Nitrogen source | ||
| 尿素 Urea | + | + |
| 硝酸钠 Sodium nitrate | + | + |
| 硝酸铵 Ammonium nitrate | + | + |
| 氯化铵 Ammonium chloride | + | + |
| 硫酸铵 Ammonium sulfate | + | + |
| 氨基酸Amino acids | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| 对照 Control | 2.57±0.21 | 106.18±2.35 |
| 谷氨酸 Glutamic acid | 2.94±0.08 | 117.15±0.56 |
| 谷氨酰胺 Glutamine | 2.69±0.18 | 106.26±1.25 |
| 脯氨酸 Proline | 2.91±0.06 | 130.46±0.56 |
表3 氨基酸添加对菌体生长和PQQ合成的影响
Table 3 Effects of amino acid addition on cell growth and PQQ synthesis
| 氨基酸Amino acids | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| 对照 Control | 2.57±0.21 | 106.18±2.35 |
| 谷氨酸 Glutamic acid | 2.94±0.08 | 117.15±0.56 |
| 谷氨酰胺 Glutamine | 2.69±0.18 | 106.26±1.25 |
| 脯氨酸 Proline | 2.91±0.06 | 130.46±0.56 |

图4 pCM66-glyA与pCM66-hps1菌液PCR鉴定 M:Trans 2k plus II DNA marker;1-2:pCM66- hps1转化子;5:pCM66-gl- yA转化子;3,6:阳性对照;4,7:阴性对照
Fig. 4 Identification of pCM66-glyA and pCM66-hps1 strain culture liquid by PCR M:Trans 2k plus II DNA marker.1-2:pCM66- hps1 transformants. 5:pCM66- glyA transformants. 3 and 6:Positive control. 4 and 7:Negative control
| 菌株Strain | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| J1-1/pCM66 | 2.63±0.15 | 82.51±1.56 |
| J1-1/pCM66-glyA | 2.86±0.18 | 99.48±2.15 |
| J1-1/pCM66- hps1 | 2.91±0.09 | 94.58±3.14 |
表4 过表达菌株表型分析
Table 4 Phenotype analysis of each overexpression strain
| 菌株Strain | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| J1-1/pCM66 | 2.63±0.15 | 82.51±1.56 |
| J1-1/pCM66-glyA | 2.86±0.18 | 99.48±2.15 |
| J1-1/pCM66- hps1 | 2.91±0.09 | 94.58±3.14 |

图6 J1-1△hps2菌株的PCR验证 M:Trans 2K plus II DNA marker;1:hps2敲除菌-Up-kan-片段PCR验证;2:J1-1菌株Up-kan-片段PCR验证;3:hps2敲除菌-kan-Down-片段PCR验证;4:J1-1菌株kan-Down-片段PCR验证;5:阴性对照
Fig.6 PCR verification of J1-1△hps2 strain M:Trans 2K plus II DNA marker. 1:pCR verification of hps2 with Up-kan-fragment knockout. 2:Up-kan-fragment PCR verification of J1-1 strain. 3:PCR verification of hps2 strain with kan-Down-fragment knowckout. 4:kan-Down-fragment PCR validation of J1-1 strain. 5:Negative control
| 菌株Strain | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| J1-1 | 3.15±0.21 | 130.45±2.35 |
| J1-1∆hps2 | 3.46±0.08 | 140.84±3.41 |
表5 hps2基因敲除菌株表型分析
Table 5 Phenotype analysis of hps2 gene knockout strain
| 菌株Strain | OD600 | PQQ/(mg·L-1) |
|---|---|---|
| J1-1 | 3.15±0.21 | 130.45±2.35 |
| J1-1∆hps2 | 3.46±0.08 | 140.84±3.41 |
| [1] |
Hauge JG. Glucose dehydrogenase of bacterium anitratum:an enzyme with a novel prosthetic group[J]. J Biol Chem, 1964, 239(11):3630-3639.
doi: 10.1016/S0021-9258(18)91183-X URL |
| [2] |
Nakano M, Kamimura A, Watanabe F, et al. Effects of orally administered pyrroloquinoline quinone disodium salt on dry skin conditions in mice and healthy female subjects[J]. J Nutr Sci Vitaminol:Tokyo, 2015, 61(3):241-246.
doi: 10.3177/jnsv.61.241 URL |
| [3] |
Shankar BS, Pandey R, Amin P, et al. Role of glutathione in augmenting the anticancer activity of pyrroloquinoline quinone(PQQ)[J]. Redox Rep, 2010, 15(4):146-154.
doi: 10.1179/174329210X12650506623762 URL |
| [4] |
Rucker R, Storms D, Sheets A, et al. Is pyrroloquinoline quinone a vitamin?[J]. Nature, 2005, 433(7025):10-11.
doi: 10.1038/433010a URL |
| [5] |
Klinman JP, Mu D. Quinoenzymes in biology[J]. Annu Rev Biochem, 1994, 63(1):299-344.
doi: 10.1146/biochem.1994.63.issue-1 URL |
| [6] | 柯崇榕. 适应性驯化选育高产吡咯喹啉醌的生丝微菌突变株[J]. 生物工程学报, 2020, 36(1):152-161. |
| Ke CR. Breeding of Hyphomicrobium denitrificans for high production of pyrroloquinoline quinone by adaptive directed domestication[J]. Chin J Biotechnol, 2020, 36(1):152-161. | |
| [7] |
Urakami T, Yashima K, Kobayashi H, et al. Production of pyrroloquinoline quinone by using methanol-utilizing bacteria[J]. Appl Environ Microbiol, 1992, 58(12):3970-3976.
doi: 10.1128/aem.58.12.3970-3976.1992 URL |
| [8] | 郑玲辉, 杜敏娜 一种生丝微菌和吡咯喹啉醌的制备方法:中国, 106282044A[P], 2015-05-20. |
| Zheng LH, Du MN. A kind of Hyphomicrobium sp. and method for the preparation of pyrroloquinoline quinone:CN, 106282044A[P], 2015-05-20. | |
| [9] | Shi W, Sun Q, Fan G, et al. gcType:a high-quality type strain genome database for microbial phylogenetic and functional research[J]. Nucleic Acids Res, 2021, 49(1):694-705. |
| [10] | Thiele I, et al. A protocol for generating a high-quality genome-scalemetabolic reconstruction[J]. Nat Protoc, 2010(1):93-121. |
| [11] | Monk JM, et al. iML1515, a knowledgebase that computes Esche-richia coli traits[J]. Nat Biotechnol, 2017(10):904-908. |
| [12] |
Brochado AR, Patil KR. Overexpression of O-methyltransferase leads to improved vanillin production in baker’s yeast only when complemented with model-guided network engineering[J]. Biotechnol Bioeng, 2013, 110(2):656-659.
doi: 10.1002/bit.24731 pmid: 23007522 |
| [13] |
Huang D, Li S, Xia M, et al. Genome-scale metabolic network guided engineering of Streptomyces tsukubaensis for FK506 production improvement[J]. Microb Cell Fact, 2013, 12:52.
doi: 10.1186/1475-2859-12-52 pmid: 23705993 |
| [14] | 王歆, 汪建华, 刘党生, 等. 吡咯喹啉醌产生菌筛选方法建立及菌种筛选[J]. 微生物学报, 2007, 47(6):982-986. |
| Wang X, Wang JH, Liu DS, et al. Establishment of the screening method and isolation of PQQ producing strains[J]. Acta Microbiol Sin, 2007, 47(6):982-986. | |
| [15] |
Ebrahim A, Lerman JA, Palsson BO, et al. COBRApy:COnstraints-based reconstruction and analysis for Python[J]. BMC Syst Biol, 2013, 7:74.
doi: 10.1186/1752-0509-7-74 pmid: 23927696 |
| [16] |
Orth JD, Thiele I, Palsson BØ. What is flux balance analysis?[J]. Nat Biotechnol, 2010, 28(3):245-248.
doi: 10.1038/nbt.1614 URL |
| [17] |
Choon YW, Mohamad MS, Deris S, et al. Differential Bees Flux Balance Analysis with OptKnock for in silico microbial strains optimization[J]. PLoS One, 2014, 9(7):e102744.
doi: 10.1371/journal.pone.0102744 URL |
| [18] |
Razmilic V, Castro JF, Andrews B, et al. Analysis of metabolic networks of Streptomyces leeuwenhoekii C34 by means of a genome scale model:Prediction of modifications that enhance the production of specialized metabolites[J]. Biotechnol Bioeng, 2018, 115(7):1815-1828.
doi: 10.1002/bit.26598 pmid: 29578590 |
| [19] | 李大攀, 等. 甲基营养菌MP688甲醇脱氢酶基因mpq1818的敲除及功能研究[J]. 生物技术通讯, 2014(5):632-635. |
| Li DP, et al. Knockout and Characterization of mpq1818 Gene of Methylovorus sp.[J]. Lett Biotechnol, 2014(5):632-635. | |
| [20] |
Xiong XH, et al. Complete genome sequence of the bacterium Methylovorus sp. strain MP688, a high-level producer of pyrroloquinolone quinone[J]. J Bacteriol, 2011, 193(4):1012-1013.
doi: 10.1128/JB.01431-10 URL |
| [21] |
Moriya Y, Itoh M, Okuda S, et al. KAAS:an automatic genome annotation and pathway reconstruction server[J]. Nucleic Acids Res, 2007, 35(web server issue):W182-W185.
doi: 10.1093/nar/gkm321 URL |
| [22] |
Kanehisa M, et al. KEGG:integrating viruses and cellular organisms[J]. Nucleic Acids Res, 2021, 49(d1):D545-D551.
doi: 10.1093/nar/gkaa970 URL |
| [23] |
King ZA, Lu J, Dräger A, et al. BiGG Models:a platform for integrating, standardizing and sharing genome-scale models[J]. Nucleic Acids Res, 2016, 44(d1):D515-D522.
doi: 10.1093/nar/gkv1049 URL |
| [24] |
Bairoch A. The ENZYME database in 2000[J]. Nucleic Acids Res, 2000, 28(1):304-305.
pmid: 10592255 |
| [25] |
Consortium U. UniProt:a worldwide hub of protein knowledge[J]. Nucleic Acids Res, 2019, 47(D1):D506-D515.
doi: 10.1093/nar/gky1049 URL |
| [26] |
Yang J, Zhang CT, Yuan XJ, et al. Metabolic engineering of Methylobacterium extorquens AM1 for the production of butadiene precursor[J]. Microb Cell Fact, 2018, 17(1):194.
doi: 10.1186/s12934-018-1042-4 URL |
| [27] |
Lapidus A, Clum A, Labutti K, et al. Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae[J]. J Bacteriol, 2011, 193(15):3757-3764.
doi: 10.1128/JB.00404-11 pmid: 21622745 |
| [28] |
Kim B, Kim WJ, Kim DI, et al. Applications of genome-scale metabolic network model in metabolic engineering[J]. J Ind Microbiol Biotechnol, 2015, 42(3):339-348.
doi: 10.1007/s10295-014-1554-9 URL |
| [29] |
Saier MH Jr, Reddy VS, et al. The transporter classification database[J]. Nucl Acids Res, 2014, 42(D1):D251-D258.
doi: 10.1093/nar/gkt1097 URL |
| [30] |
Feist AM, Palsson BØ. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli[J]. Nat Biotechnol, 2008, 26(6):659-667.
doi: 10.1038/nbt1401 pmid: 18536691 |
| [31] |
Latendresse M. Efficiently gap-filling reaction networks[J]. BMC Bioinformatics, 2014, 15:225.
doi: 10.1186/1471-2105-15-225 pmid: 24972703 |
| [32] |
Ge X, Wang W, Du B, et al. Multiple pqqA genes respond differently to environment and one contributes dominantly to pyrroloquinoline quinone synjournal[J]. J Basic Microbiol, 2015, 55(3):312-323.
doi: 10.1002/jobm.v55.3 URL |
| [33] |
Kim J, Coradetti ST, Kim YM, et al. Multi-omics driven metabolic network reconstruction and analysis of lignocellulosic carbon utilization in Rhodosporidium toruloides[J]. Front Bioeng Biotechnol, 2020, 8:612832.
doi: 10.3389/fbioe.2020.612832 URL |
| [34] |
Müller JE, Litsanov B, Bortfeld-Miller M, et al. Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3[J]. Proteomics, 2014, 14(6):725-737.
doi: 10.1002/pmic.201300515 pmid: 24452867 |
| [35] |
Ochsner AM, Sonntag F, Buchhaupt M, et al. Methylobacterium extorquens:methylotrophy and biotechnological applications[J]. Appl Microbiol Biotechnol, 2015, 99(2):517-534.
doi: 10.1007/s00253-014-6240-3 pmid: 25432674 |
| [36] | van Dien SJ, et al. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobac-terium extorquens AM1, with application to reconstruction of C3 and C4 metabolism[J]. Biotechnol Bioeng, 2002(3):296-312. |
| [37] |
Peyraud R, Schneider K, et al. Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1[J]. BMC Syst Biol, 2011, 5:189.
doi: 10.1186/1752-0509-5-189 pmid: 22074569 |
| [1] | 郑焕, 林冬梅, 刘峻源, 张引莲, 林标声, 林占熺, 李晶. 基于LC-QTOF-MS代谢组学解析牛樟芝子实体和菌丝体中氨基酸代谢差异[J]. 生物技术通报, 2023, 39(5): 254-266. |
| [2] | 艾露, 陈文慧, 史京辉, 任志远, 沈文琦, 杨嘉凝, 骆健美, 王敏. 赭曲霉11α羟化酶的克隆表达及关键氨基酸位点分析[J]. 生物技术通报, 2023, 39(4): 114-123. |
| [3] | 王小琴, 黄银萍, 王蔚倩, 吴萍, 全舒. 含非天然氨基酸定点突变的MLL3SET蛋白表达与纯化[J]. 生物技术通报, 2022, 38(3): 194-202. |
| [4] | 任思羽, 程新宽, 张宇辉, 庄建文, 马龙. 两种新型L-苏氨酸醛缩酶的鉴定及活性检测方法[J]. 生物技术通报, 2021, 37(3): 233-240. |
| [5] | 郝俊尧, 马富强, 杨广宇. 产碱杆菌Alcaligenes sp.KS-85来源肌酸酶活性中心的关键氨基酸功能研究[J]. 生物技术通报, 2021, 37(3): 75-83. |
| [6] | 郑惠清, 郭仲杰, 蔡志欣, 卢园萍, 廖剑华, 陈美元. 双孢蘑菇野生种质资源营养成分分析与评价[J]. 生物技术通报, 2021, 37(11): 109-118. |
| [7] | 郑博, 王宁, 霍毅欣. 基于转录和翻译调控的氨基酸高产菌株筛选及构建策略[J]. 生物技术通报, 2020, 36(4): 34-40. |
| [8] | 赵杰, 岳华, 苟学磊, 周金燕, 谭红. 伊枯草菌素A发酵过程中游离氨基酸的HPLC分析[J]. 生物技术通报, 2018, 34(8): 151-158. |
| [9] | 马林龙, 曹丹, 刘艳丽, 金孝芳. 组学技术在高氨基酸茶树中的应用研究进展[J]. 生物技术通报, 2018, 34(3): 39-42. |
| [10] | 侯若琳, 刘鑫, 项凯凯, 陈磊, 郑明锋, 傅俊生. 响应面法优化蛹虫草蛋白超声波辅助提取工艺及其营养评价[J]. 生物技术通报, 2018, 34(11): 198-204. |
| [11] | 胡冬冬, 赵亮, 范里, 刘旭平, 邓献存, 缪仕伟, 谭文松. 酵母抽提物对CHO细胞生长及抗体表达的影响[J]. 生物技术通报, 2017, 33(6): 162-169. |
| [12] | 易华伟唐晓峰. 基于氨基酸序列和模拟结构预测蛋白质稳定性的研究进展[J]. 生物技术通报, 2017, 33(4): 83-89. |
| [13] | 申晓林, 袁其朋. 生物合成芳香族氨基酸及其衍生物的研究进展[J]. 生物技术通报, 2017, 33(1): 24-34. |
| [14] | 王琪, 齐仁立, 王敬, 黄金秀. 氨基酸缺乏诱导细胞自噬及miRNA调控机制[J]. 生物技术通报, 2016, 32(9): 38-43. |
| [15] | 丛华剑,武栓虎,田健,初晓宇,伍宁丰. 基于细菌同源蛋白预测细菌最适生长温度的研究[J]. 生物技术通报, 2016, 32(3): 155-165. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||