








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 194-202.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0465
王小琴1( ), 黄银萍1, 王蔚倩2, 吴萍2, 全舒1(
), 黄银萍1, 王蔚倩2, 吴萍2, 全舒1( )
)
收稿日期:2021-04-09
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:王小琴,女,博士研究生,研究方向:酶的催化机制;E-mail: 基金资助:
WANG Xiao-qin1( ), HUANG Yin-ping1, WANG Wei-qian2, WU Ping2, QUAN Shu1(
), HUANG Yin-ping1, WANG Wei-qian2, WU Ping2, QUAN Shu1( )
)
Received:2021-04-09
Published:2022-03-26
Online:2022-04-06
摘要:
在组蛋白H3K4甲基转移酶MLL3的催化结构域(MLL3SET)中定点引入非天然氨基酸N-炔丙基赖氨酸(N-propargyl-lysine,PrK),表达、纯化该突变蛋白(MLL3SET*),并评估突变蛋白的酶活,为后续进一步利用单分子荧光共振能量转移技术(smFRET)表征MLL3的作用机制奠定基础。将MLL3SET接入pET-28b(+)构建表达载体,通过MLL3SET晶体结构分析选择N4905位点进行PrK的引入;对商业化菌株(C321. ΔA. exp)进行基因组改造以引入T7 RNA聚合酶基因,并在改造后的菌株中表达、纯化MLL3SET*,最后测定MLL3SET*的酶活性。结果表明,在构建的C321. ΔA. exp lacZ∷T7p07菌株中,pET28b-MLL3SET* 在共转入aaRS-tRNA正交系统以及外源添加PrK后能够正常表达;通过Ni-NTA亲和层析及凝胶过滤层析成功纯化出高纯度的MLL3SET*蛋白;多酶级联反应结果显示,MLL3SET*的酶活性比野生型蛋白低,但仍具有约43%的甲基转移酶活性。本研究成功实现了含PrK的MLL3SET 蛋白的原核表达与纯化,突变蛋白保留了一定酶活性,为后续深入研究MLL3的分子机制奠定了基础。
王小琴, 黄银萍, 王蔚倩, 吴萍, 全舒. 含非天然氨基酸定点突变的MLL3SET蛋白表达与纯化[J]. 生物技术通报, 2022, 38(3): 194-202.
WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid[J]. Biotechnology Bulletin, 2022, 38(3): 194-202.
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Kan-F | TTCGCGTTCGCGTAAGTGTAGGCTGGAGCTGC |
| Kan-R | TTACGCGAAATACGGGCAGACATGGCCTGCC- CGGTTATTAATATCCTCCTTAGTTCCTATTCC |
| T7p07-F | GGAATTGTGAGCGGATAACAATTTCACACAGG- AAACAGCTATGAACACGATTAACATCGCTAAG |
| T7p07-R | CTCCAGCCTACACTTACGCGAACGCGAAGTCC |
| (T7p07+Kan)-F | GGAATTGTGAGCGGATAAC |
| (T7p07+Kan)-R | TTACGCGAAATACGGGCAG |
| F1 | CCAGAAAGGAGAAGAGCTCTCCTATGACTAT- AAGTTTGAC |
| R1 | GTCAAACTTA TAGTCATAG GAGAGCTCTTCT- CCTTTCTGG |
| F2 | GAGAAGCTTTATGAGTGTCAGAACCGTGGTG- TGTAC |
| R2 | GTACACACCACGGTTCTGACACTCATAAAGC- TTCTC |
| F3 | GTGGAGCTGTGTAGTGCCGGAAGTGGATG |
| R3 | CATCCACTTCCGGCACTACACAGCTCCAC |
表1 PCR反应引物序列
Table 1 Primer sequences of PCR reactions
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Kan-F | TTCGCGTTCGCGTAAGTGTAGGCTGGAGCTGC |
| Kan-R | TTACGCGAAATACGGGCAGACATGGCCTGCC- CGGTTATTAATATCCTCCTTAGTTCCTATTCC |
| T7p07-F | GGAATTGTGAGCGGATAACAATTTCACACAGG- AAACAGCTATGAACACGATTAACATCGCTAAG |
| T7p07-R | CTCCAGCCTACACTTACGCGAACGCGAAGTCC |
| (T7p07+Kan)-F | GGAATTGTGAGCGGATAAC |
| (T7p07+Kan)-R | TTACGCGAAATACGGGCAG |
| F1 | CCAGAAAGGAGAAGAGCTCTCCTATGACTAT- AAGTTTGAC |
| R1 | GTCAAACTTA TAGTCATAG GAGAGCTCTTCT- CCTTTCTGG |
| F2 | GAGAAGCTTTATGAGTGTCAGAACCGTGGTG- TGTAC |
| R2 | GTACACACCACGGTTCTGACACTCATAAAGC- TTCTC |
| F3 | GTGGAGCTGTGTAGTGCCGGAAGTGGATG |
| R3 | CATCCACTTCCGGCACTACACAGCTCCAC |
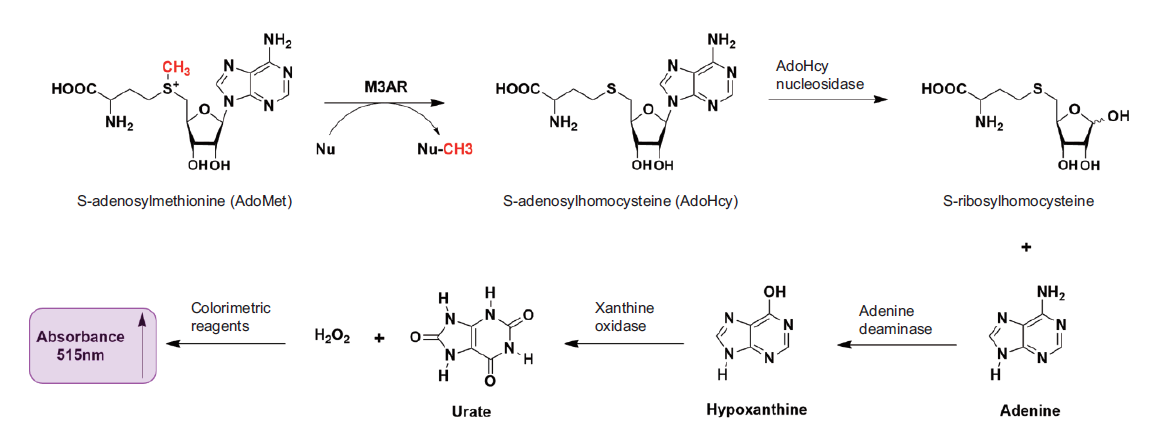
图1 多酶级联反应测定MLL3SET*活性的原理图 在MLL3SET和AR复合物(M3AR)及其他酶的催化下,SAM经过多步反应,最终生成的物质N-(4-安替比林)-3-氯-5-磺酸酯苯醌-单亚胺在515 nm处具有吸光度
Fig. 1 Diagram of the multi-enzyme coupling reaction for determining MLL3SET* activity Under the catalysis of MLL3SET and AR compound(M3AR)and other enzymes,SAM undergoes multiple reactions. The final product N-(4-antipyryl)-3-chloro-5-sulfonate-ρ-benzoquinone-monoimine shows absorbance at 515 nm
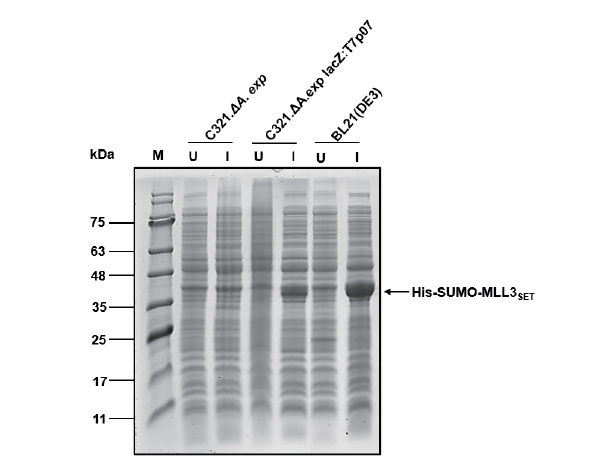
图2 基因组改造表达测试 M:蛋白分子量标准;U:未诱导总菌体裂解液;I:诱导总菌体裂解液
Fig. 2 Expression test after genomic modification M:Protein marker. U:Lysate of uninduced whole cell. I:Lysate of induced whole cell

图3 MLL3SET结构信息(A)和表达质粒图谱(B) 将MLL3SET的第4 883位半胱氨酸突变为丝氨酸,将4 819位丝氨酸突变为半胱氨酸,将4 905位的天冬酰胺突变为琥珀密码子。A:MLL3SET六个半胱氨酸的位置信息(PDB:5F6K),MLL3SET结构为黄色;底物H3为海蓝色;产物SAH、6个半胱氨酸以及2个标记位点用棍棒模型显示;锌离子用灰色小球显示;B:pET28b-His-sumo-mll3set C4883S S4819C N4905X质粒图谱
Fig. 3 MLL3SET structural information(A)and the map of the expression plasmid (B) The cysteine at position 4 883 of MLL3SET is mutated to serine,serine at position 4 819 to cysteine,and asparagine at position 4 905 to the amber codon. A:Structural information of 6 cysteine residues in MLL3SET(PDB:5F6K). MLL3SET is colored in yellow;the substrate H3 is colored in aquamarine. The product,SAH,6 cysteines,and the two sites chosen for smFRET labeling are displayed as the stick model. The zinc ion is shown in the grey sphere. B:pET28b-His-sumo-mll3set C4883S S4819C N4905X plasmid map

图4 pET28b-His-sumo-mll3set C4883S质粒的表达测试 M:蛋白分子量标准;U:未诱导总菌体裂解液;I:诱导总菌体裂解液;S:诱导裂解液上清;P:诱导裂解液沉淀;37℃:在37℃表达;16℃:在16℃表达
Fig. 4 Expression test of the pET28b-His-sumo-mll3set C4883S plasmid M:Protein marker. U:Uninduced whole cell after lysis. I:Induced whole cell after lysis. S:Induced supernatant after lysis. P:Induced precipitation after lysis. 37℃:Expression at 37℃. 16℃:Expression at 16℃
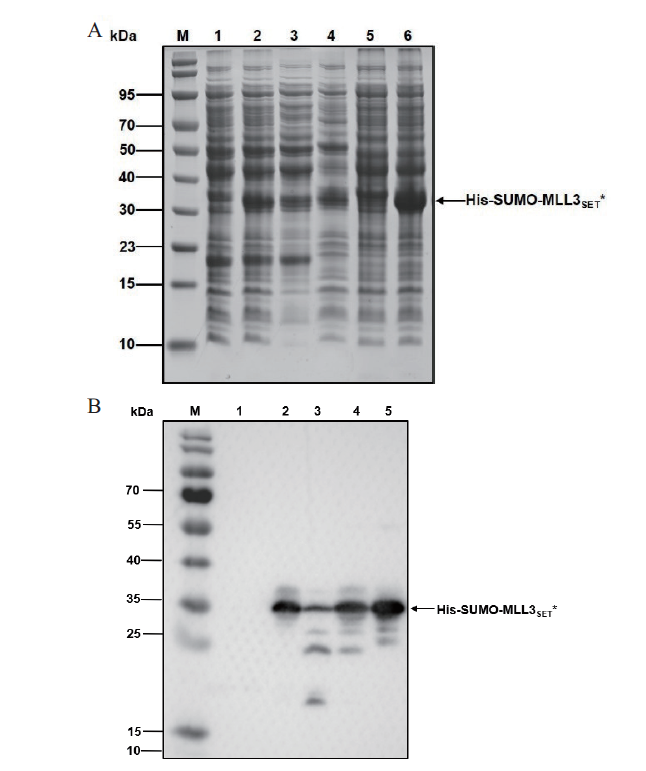
图5 表达His-SUMO-MLL3SET*分析 A:SDS-PAGE分析;B:Western blot分析. M:蛋白分子质量标准;1-4:宿主菌为C321.ΔA. exp lacZ∷T7;1:未诱导总菌体裂解液;2:诱导总菌体裂解液;3:诱导裂解液上清;4:诱导裂解液沉淀;5-6:宿主菌为BL21(DE3);5:未诱导总菌体裂解液;6:诱导总菌体裂解液
Fig. 5 Analysis of expressed His-SUMO-MLL3SET* A:SDS-PAGE analysis. B:Western blot analysis. M:Protein marker;1-4:The host strain was E. coli C321.ΔA. exp lacZ∷T7. 1:Uninduced whole cell after lysis. 2:Induced whole cell after lysis. 3:Supernatant of induced cells after lysis. 4:Precipitation of induced cells after lysis. 5-6:The host strain was E. coli BL21(DE3). 5:Uninduced whole cell after lysis. 6:Induced whole cell after lysis

图6 纯化His-SUMO-MLL3SET*分析 M:蛋白分子质量标准;1:诱导总菌体裂解液;2:诱导裂解液上清;3:诱导裂解液沉淀;4:洗涤前的流出缓冲液;5:用50 mL 缓冲液洗涤后的漂洗液;6:用1 L洗涤缓冲液洗涤后的漂洗液;7:用缓冲液重悬混合均匀咪唑洗脱前的Ni beads;8:咪唑洗脱液;9:用缓冲液重悬混合均匀咪唑洗脱后的Ni beads
Fig. 6 Analysis of purified His-SUMO-MLL3SET* M:Protein marker. 1:Induced whole cell. 2:Supernatant of the induced cells. 3:Precipitation of the induced cells. 4:Flow through before washing. 5:Flow through after washing with 50 mL buffer. 6:Flow through after washing with 1 L buffer. 7:Suspended Ni beads with buffer before imidazole elution. 8:Imidazole eluted sample. 9:Suspended Ni beads with buffer after imidazole elution

图9 多酶级联反应测His-SUMO-MLL3SET*活性 A:M*AR与MAR活性比较;B:SAH浓度与A515的标准曲线
Fig. 9 Activity of His-SUMO-MLL3SET* measured by a multi-enzyme coupling reaction A:Comparison of the activity of M*AR and MAR. B:Standard curve of SAH concentration and A515
| [1] |
Shilatifard A. The COMPASS family of histone H3K4 methylases:mechanisms of regulation in development and disease pathogenesis[J]. Annu Rev Biochem, 2012, 81:65-95.
doi: 10.1146/annurev-biochem-051710-134100 pmid: 22663077 |
| [2] |
Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins[J]. Oncogene, 2001, 20(40):5695-5707.
pmid: 11607819 |
| [3] | 黎彦璟, 陈勇. 组蛋白甲基转移酶MLL1的结构与功能研究进展[J]. 中国细胞生物学学报, 2014, 36(7):857-868. |
| Li YJ, Chen Y. Progress in structural and functional studies of histone methyltransferase MLL1[J]. Chin J Cell Biol, 2014, 36(7):857-868. | |
| [4] |
Dehé PM, Dichtl B, Schaft D, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation[J]. J Biol Chem, 2006, 281(46):35404-35412.
doi: 10.1074/jbc.M603099200 URL |
| [5] |
Avdic V, Zhang P, Lanouette S, et al. Structural and biochemical insights into MLL1 core complex assembly[J]. Structure, 2011, 19(1):101-108.
doi: 10.1016/j.str.2010.09.022 URL |
| [6] |
Zhang Y, Mittal A, Reid J, et al. Evolving catalytic properties of the MLL family SET domain[J]. Structure, 2015, 23(10):1921-1933.
doi: S0969-2126(15)00324-X pmid: 26320581 |
| [7] |
Southall SM, Wong PS, Odho Z, et al. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks[J]. Mol Cell, 2009, 33(2):181-191.
doi: 10.1016/j.molcel.2008.12.029 pmid: 19187761 |
| [8] |
Li Y, Han J, Zhang Y, et al. Structural basis for activity regulation of MLL family methyltransferases[J]. Nature, 2016, 530(7591):447-452.
doi: 10.1038/nature16952 URL |
| [9] |
Sasmal DK, Pulido LE, Kasal S, et al. Single-molecule fluorescence resonance energy transfer in molecular biology[J]. Nanoscale, 2016, 8(48):19928-19944.
doi: 10.1039/C6NR06794H URL |
| [10] | Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET[J]. Nat Methods, 2008, 5(6):507-516. |
| [11] |
Zhang J, Fu Y, Lakowicz JR. Enhanced förster resonance energy transfer(FRET)on single metal particle[J]. J Phys Chem C Nanomater Interfaces, 2007, 111(1):50-56.
doi: 10.1021/jp062665e URL |
| [12] |
Zhang J, Fu Y, Chowdhury MH, et al. Enhanced förster resonance energy transfer on single metal particle. 2. dependence on donor-acceptor separation distance, particle size, and distance from metal surface[J]. J Phys Chem C, 2007, 111(32):11784-11792.
doi: 10.1021/jp067887r pmid: 19890406 |
| [13] |
Lee TC, Kang M, Kim CH, et al. Dual unnatural amino acid incorporation and click-chemistry labeling to enable single-molecule FRET studies of p97 folding[J]. Chembiochem, 2016, 17(11):981-984.
doi: 10.1002/cbic.v17.11 URL |
| [14] |
Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins[J]. Chem Rev, 2014, 114(9):4764-4806.
doi: 10.1021/cr400355w URL |
| [15] | Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli[J]. Nat Methods, 2006, 3(4):263-265. |
| [16] |
Young TS, Ahmad I, Yin JA, et al. An enhanced system for unnatural amino acid mutagenesis in E. coli[J]. J Mol Biol, 2010, 395(2):361-374.
doi: 10.1016/j.jmb.2009.10.030 URL |
| [17] |
Smolskaya S, Andreev Y. Site-specific incorporation of unnatural amino acids into Escherichia coli recombinant protein:methodology development and recent achievement[J]. Biomolecules, 2019, 9(7):255.
doi: 10.3390/biom9070255 URL |
| [18] |
Lin X, Yu ACS, Chan TF. Efforts and challenges in engineering the genetic code[J]. Life, 2017, 7(1):12.
doi: 10.3390/life7010012 URL |
| [19] |
Young DD, Schultz PG. Playing with the molecules of life[J]. ACS Chem Biol, 2018, 13(4):854-870.
doi: 10.1021/acschembio.7b00974 URL |
| [20] |
Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases[J]. Anal Biochem, 2006, 350(2):249-255.
doi: 10.1016/j.ab.2006.01.004 URL |
| [21] | Liu ZP. Histone methylation in heart development and cardiovascular disease[J]. Cardiac and Vascular Biology, 2016, 1:125-146. |
| [22] |
Bale TL, Baram TZ, Brown AS, et al. Early life programming and neurodevelopmental disorders[J]. Biol Psychiatry, 2010, 68(4):314-319.
doi: 10.1016/j.biopsych.2010.05.028 URL |
| [23] |
Millan MJ. An epigenetic framework for neurodevelopmental disorders:from pathogenesis to potential therapy[J]. Neuropharmacology, 2013, 68:2-82.
doi: 10.1016/j.neuropharm.2012.11.015 pmid: 23246909 |
| [24] |
Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders[J]. Nat Rev Genet, 2016, 17(1):9-18.
doi: 10.1038/nrg3999 URL |
| [1] | 赵忠娟, 杨凯, 扈进冬, 魏艳丽, 李玲, 徐维生, 李纪顺. 盐胁迫条件下哈茨木霉ST02对椒样薄荷生长及根区土壤理化性质的影响[J]. 生物技术通报, 2022, 38(7): 224-235. |
| [2] | 贾海红, 李冰清. 超氧化物歧化酶翻译后修饰的研究进展[J]. 生物技术通报, 2022, 38(2): 237-244. |
| [3] | 武杞蔓, 田诗涵, 李昀烨, 潘英杰, 张颖. 微生物菌肥对设施黄瓜生长、产量及品质的影响[J]. 生物技术通报, 2022, 38(1): 125-131. |
| [4] | 曹汝菲, 李泽轩, 许欢, 张莎, 张敏敏, 戴枫, 段晓雷. 脆弱拟杆菌Pif1解旋酶的表达纯化与晶体生长[J]. 生物技术通报, 2021, 37(9): 180-190. |
| [5] | 陈晓雨, 张建, 张新亚, 唐雨婷, 邵钰晨, 罗志丹, 卢辰. 一种快速精确测定Tth DNA聚合酶活性的方法[J]. 生物技术通报, 2021, 37(5): 281-286. |
| [6] | 谢果珍, 唐圆, 吴仪, 黄莉莉, 谭周进. 七味白术散总苷对菌群失调腹泻小鼠肠道微生物及酶活性的影响[J]. 生物技术通报, 2021, 37(12): 124-131. |
| [7] | 田庚, 高伟强, 陈晓波, 张春晓. 地衣芽孢杆菌KD-1β-甘露聚糖酶定点突变提高酶活性及稳定性[J]. 生物技术通报, 2021, 37(10): 100-109. |
| [8] | 袁亮. 微生物碳酸酐酶诱导CaCO3沉淀的影响因素及生成机理[J]. 生物技术通报, 2020, 36(8): 79-68. |
| [9] | 殷金瑶, 王义, 徐良向, 朱利, 王晨, 刘文波, 缪卫国. 橡胶树白粉菌(HO-73)启动子WY172不同长度片段的克隆及表达活性分析[J]. 生物技术通报, 2020, 36(1): 29-36. |
| [10] | 胡楚霄, 雷善钰, 秦艳平, 赵奕锦, 向泉桔. 蒽对3株灵芝菌株漆酶活性及其转录表达水平的影响[J]. 生物技术通报, 2019, 35(9): 112-117. |
| [11] | 岳鑫, 杨爱江, 徐鹏, 胡霞, 朱桓毅, 包欣. 锑胁迫对斑马鱼酶活性的影响研究[J]. 生物技术通报, 2019, 35(6): 99-106. |
| [12] | 郭珺, 樊芳芳, 王立革, 武爱莲, 郑军. 固碳微生物菌株的分离鉴定及其固碳能力测定[J]. 生物技术通报, 2019, 35(1): 90-97. |
| [13] | 秦海彬, 熊涛, 张博, 牛坤. α-酮戊二酸半醛脱氢酶的定点突变及酶学性质变化 [J]. 生物技术通报, 2017, 33(8): 180-185. |
| [14] | 孙雯, 郑峰. S. suis 2 中国强毒株烯醇化酶 Enolase 基因的分子克隆及蛋白生物功能研究[J]. 生物技术通报, 2017, 33(4): 222-230. |
| [15] | 刘小利, 江世杰, 薛冬, 刘盈盈, 吴小丽, 冯帅, 韩佳慧, 汪雨舟, 平淑珍, 王劲. 耐辐射异常球菌dlp基因缺失突变株构建及其生物学功能研究[J]. 生物技术通报, 2017, 33(2): 155-163. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||