








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 173-180.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0647
燕炯1( ), 冯晨毅1, 高学坤1, 许祥1, 杨佳敏1, 陈朝阳2(
), 冯晨毅1, 高学坤1, 许祥1, 杨佳敏1, 陈朝阳2( )
)
收稿日期:2021-05-19
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:燕炯,男,博士,副教授,研究方向:营养相关疾病分子学;E-mail: 基金资助:
YAN Jiong1( ), FENG Chen-yi1, GAO Xue-kun1, XU Xiang1, YANG Jia-min1, CHEN Zhao-yang2(
), FENG Chen-yi1, GAO Xue-kun1, XU Xiang1, YANG Jia-min1, CHEN Zhao-yang2( )
)
Received:2021-05-19
Published:2022-03-26
Online:2022-04-06
摘要:
利用CRISPR/Cas9技术构建纯合围脂滴蛋白(Plin1)基因敲除小鼠模型,并初步分析其表型。针对Plin1基因2号外显子前后序列设计sgRNA并构建表达载体,体外转录获得sgRNA后与Cas9蛋白混合,显微注射至小鼠受精卵中并进行胚胎移植。出生小鼠经测序及PCR基因型鉴定获得F0代阳性小鼠;令F0代小鼠与野生型小鼠杂交,获得F1代杂合子小鼠;通过F1代杂合小鼠近交,获得F2代纯合子小鼠模型。常规饲喂同窝别Plin1基因敲除纯合小鼠、杂合小鼠和野生型小鼠,并测量小鼠体重、身长等体格参数;定量实时聚合酶链反应(Q-RT-PCR)和蛋白质印迹(WB)分别检测每个组织中Plin1基因在mRNA和蛋白质水平的表达。敲除了小鼠Plin1基因741 bp的片段(包含了2号外显子在内);Plin1基因敲除小鼠PLIN1 mRNA和蛋白表达水平均显著降低(P < 0.05);常规喂养4周后,相比于野生型及杂合型小鼠,纯合型Plin1基因敲除小鼠体重显著降低(P < 0.05)。成功构建了Plin1基因敲除小鼠模型;初步表型分析发现,Plin1基因敲除小鼠与野生型小鼠相比较为瘦弱。
燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180.
YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology[J]. Biotechnology Bulletin, 2022, 38(3): 173-180.
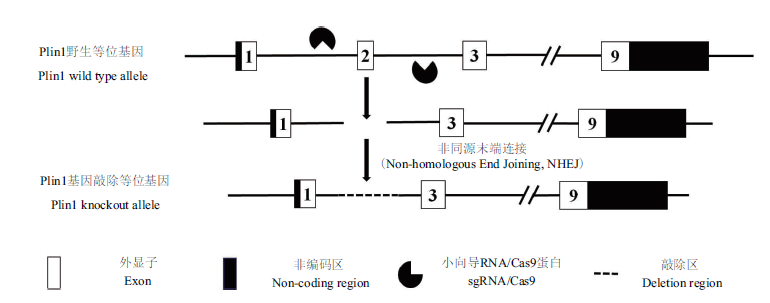
图1 sgRNA敲除Plin1基因2号外显子的示意图 Plin1基因转录本为NM_001113471.1
Fig. 1 Schematic diagram of sgRNA knocking out exon 2 of Plin1 gene The Plin1 gene transcript is NM_001113471.1
| 名称 Name | 序列 Sequence(5'-3') | PAM序列 PAM sequence |
|---|---|---|
| sgRNA-Plin1-1 | TTGGAGGGCGAATGTTACAT | AGG |
| sgRNA-Plin1-2 | GAACTTCAGGCGTATGACCC | TGG |
表1 sgRNA序列
Table 1 sgRNA sequence
| 名称 Name | 序列 Sequence(5'-3') | PAM序列 PAM sequence |
|---|---|---|
| sgRNA-Plin1-1 | TTGGAGGGCGAATGTTACAT | AGG |
| sgRNA-Plin1-2 | GAACTTCAGGCGTATGACCC | TGG |
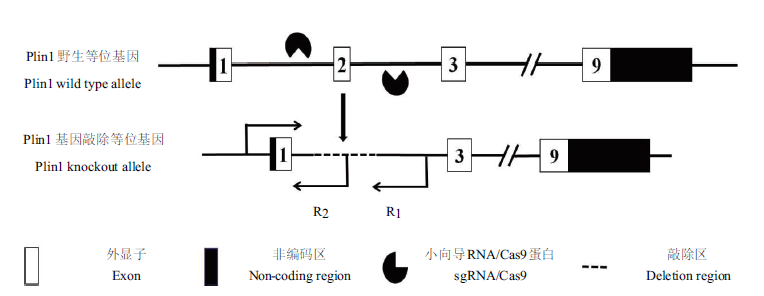
图2 Plin1基因小鼠敲除基因型鉴定的策略 Plin1基因转录本为NM_001113471.1。
Fig. 2 Strategies for genotype identification of Plin1 knoc-kout mice The Plin1 gene transcript is NM_001113471.1
| 名称Name | 序列Sequence(5'-3') | 大小Size/bp |
|---|---|---|
| Plin1-F | CTGAGAGAAGGCTTAACCTTGCTGG | 25 |
| Plin1-R1 | AGCTTTCCATCCTGCAAGTGAGTCAG | 26 |
| Plin1-R2 | AGGTGGCAAGGACAGAGACAGTGAG | 25 |
表2 基因型鉴定的引物序列
Table 2 Primer sequence for genotyping
| 名称Name | 序列Sequence(5'-3') | 大小Size/bp |
|---|---|---|
| Plin1-F | CTGAGAGAAGGCTTAACCTTGCTGG | 25 |
| Plin1-R1 | AGCTTTCCATCCTGCAAGTGAGTCAG | 26 |
| Plin1-R2 | AGGTGGCAAGGACAGAGACAGTGAG | 25 |
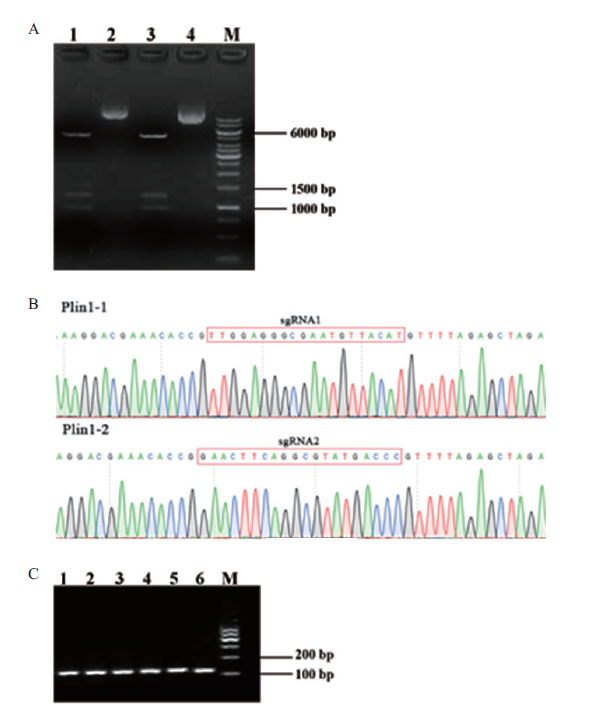
图3 sgRNA表达载体构建与鉴定 A:sgRNA表达载体的酶切结果(2,4泳道:sgRNA-Plin1-1、2载体;1,3泳道:酶切片段;M:DNA marker);B:sgRNA表达载体测序结果;C:sgRNA体外转录片段电泳结果(1-3泳道:sgRNA-Plin1-1转录产物;4-6:sgRNA-Plin1-2转录产物;M:DNA marker)
Fig. 3 Construction and identification of sgRNA expression vector A:Digestion results of sgRNA expression vector(Lane 2 and 4:sgRNA-Plin1-1,2 vectors. Lane 1 and 3:Enzyme fragment. M:DNA marker). B:Sequencing results of sgRNA expression vector. C:In vitro transcriptional fragment electrophoresis results of sgRNA(Lane 1-3:sgRNA-Plin1-1 transcription products. Lane 4-6 lane:sgRNA-Plin1-2 transcription products. M:DNA marker)
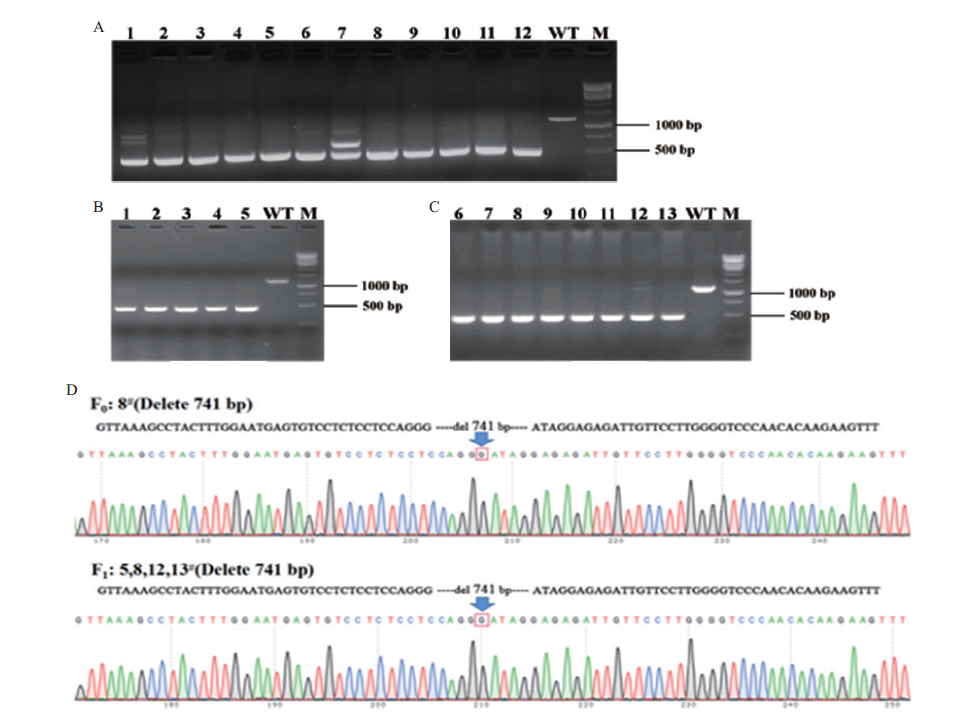
图4 F0和F1代小鼠基因型鉴定 A:F0代阳性小鼠Plin1-F1/R1扩增Plin1基因的电泳结果;1-12泳道:F0阳性小鼠;WT:野生型对照;M:DNA marker;B-C:F1代小鼠Plin1-F1/R1扩增Plin1基因的电泳结果。1-13泳道:F1代小鼠;WT:野生型对照;M:DNA marker;D:F0、F1代小鼠基因测序结果
Fig. 4 Genotype identification of F0 and F1 generation mice A:Electrophoretic results of Plin1-F1/R1 amplified from F0 generation positive mice;lane 1-12:F0 positive mice;WT:wild type control;M. DNA marker. B-C:Electrophoretic results of Plin1-F1/R1 amplified from F1 generation mice;lane 1-13:F1 generation mice;WT:wild type control. M:DNA marker. D:gene sequencing results of F0 and F1 generation mice
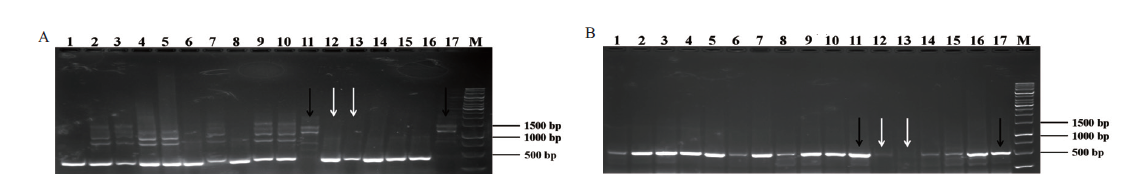
图5 F2代小鼠基因型鉴定 A:Plin1-F1/R1扩增Plin1基因的电泳结果;B:Plin1-F1/R2扩增Plin1基因的电泳结果;1-17泳道:F2代小鼠Plin1基因片段;白色箭头:纯合型;黑色箭头:野生型
Fig. 5 Genotype identification of F2 generation mice A:Electrophoretic results of Plin1 gene amplified by Plin1-F1/R1. B:Electrophoretic results of Plin1 gene amplified by Plin1-F1/R2. Lane 1-17:Plin1 gene fragment of F2 generation mice. White arrow:Homozygous. Black arrow:Wild type
| 组别/指标 Group/index | 体重 Body weight/g | 体长 Body length/cm | 摄食量 Food intake/g |
|---|---|---|---|
| 野生型WT | 22.95±0.55 | 8.88±2.17 | 16.26±2.13 |
| 杂合型Plin1+/- | 23.20±1.57 | 9.31±3.49 | 15.43±3.03 |
| 纯合型Plin1-/- | 18.34±0.35* | 8.72±1.34 | 15.67±2.59 |
表3 各组小鼠体格参数
Table 3 Physical parameters of mice in each group
| 组别/指标 Group/index | 体重 Body weight/g | 体长 Body length/cm | 摄食量 Food intake/g |
|---|---|---|---|
| 野生型WT | 22.95±0.55 | 8.88±2.17 | 16.26±2.13 |
| 杂合型Plin1+/- | 23.20±1.57 | 9.31±3.49 | 15.43±3.03 |
| 纯合型Plin1-/- | 18.34±0.35* | 8.72±1.34 | 15.67±2.59 |
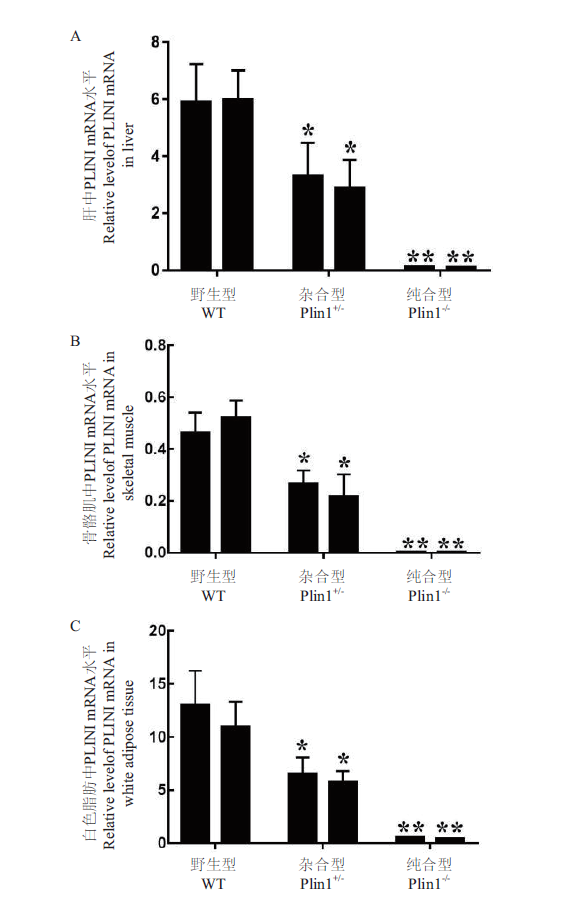
图6 小鼠各组织中PLIN1 mRNA的相对表达量 A:肝脏中PLIN1 mRNA相对表达量;B:骨骼肌中PLIN1 mRNA相对表达量;C:白色脂肪组织中PLIN1 mRNA相对表达量;**:相比于Plin1+/-,P < 0.05;*:相比于WT,P < 0.05,下同
Fig. 6 Relative expressions of PLIN1 mRNA in the various tissues of mice A:Relative level of PLINI mRNA in liver. B:Relative level of PLINI mRNA in skeletal muscle. C:Relative level of PLINI mRNA in white adipose tissue. **:Com-pared with Plin1+/-. P < 0.05. *:Compared with WT,P < 0.05. The same below
| [1] |
Jarc E, Petan T. Lipid droplets and the management of cellular stress[J]. Yale J Biol Med, 2019, 92(3):435-452.
pmid: 31543707 |
| [2] |
Jackson CL. Lipid droplet biogenesis[J]. Curr Opin Cell Biol, 2019, 59:88-96.
doi: 10.1016/j.ceb.2019.03.018 URL |
| [3] |
Itabe H, Yamaguchi T, Nimura S, et al. Perilipins:a diversity of intracellular lipid droplet proteins[J]. Lipids Health Dis, 2017, 16(1):83.
doi: 10.1186/s12944-017-0473-y URL |
| [4] |
Huang X, Sun J, Bian C, et al. Perilipin 1-3 in grass carp Ctenopharyngodon idella:molecular characterization, gene structure, tissue distribution, and mRNA expression in DHA-induced lipid droplet formation in adipocytes[J]. Fish Physiol Biochem, 2020, 46(6):2311-2322.
doi: 10.1007/s10695-020-00857-x URL |
| [5] |
Westhoff CC, Mrozinski J, Riedel I, et al. Perilipin 1 is a highly specific marker for adipocytic differentiation in sarcomas with intermediate sensitivity[J]. J Cancer Res Clin Oncol, 2017, 143(2):225-232.
doi: 10.1007/s00432-016-2263-8 URL |
| [6] |
Zhang S, Liu G, Xu C, et al. Perilipin 1 mediates lipid metabolism homeostasis and inhibits inflammatory cytokine synjournal in bovine adipocytes[J]. Front Immunol, 2018, 9:467.
doi: 10.3389/fimmu.2018.00467 URL |
| [7] |
Sohn JH, Lee YK, Han JS, et al. Perilipin 1(Plin1)deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation[J]. J Biol Chem, 2018, 293(36):13974-13988.
doi: 10.1074/jbc.RA118.003541 URL |
| [8] |
de Oliveira BAP, de Souza Pinhel MA, Nicoletti CF, et al. UCP2 and PLIN1 expression affects the resting metabolic rate and weight loss on obese patients[J]. Obes Surg, 2017, 27(2):343-348.
doi: 10.1007/s11695-016-2275-0 pmid: 27376365 |
| [9] | Zou L, Wang W, Liu S, et al. Spontaneous hypertension occurs with adipose tissue dysfunction in perilipin-1 null mice[J]. Biochim Biophys Acta, 2016, 1862(2):182-191. |
| [10] |
Mori Y, Otabe S, Dina C, et al. Genome-wide search for type 2 diabetes in Japanese affected sib-pairs confirms susceptibility genes on 3q, 15q, and 20q and identifies two new candidate Loci on 7p and 11p[J]. Diabetes, 2002, 51(4):1247-1255.
doi: 10.2337/diabetes.51.4.1247 URL |
| [11] | 陈燕波, 常翠青, 黄志卓, 等. PLIN基因多态性在中国汉族成年肥胖者中的分布[J]. 营养学报, 2011, 33(1):29-33. |
| Chen YB, Chang CQ, Huang ZZ, et al. Distribution of plin gene polymorphism in Chinese Han obese adults[J]. Acta Nutr Sin, 2011, 33(1):29-33. | |
| [12] | Hryhorowicz M, Lipiński D, Zeyland J, et al. CRISPR/Cas9 immune system as a tool for genome engineering[J]. Arch Immunol Ther Exp:Warsz, 2017, 65(3):233-240. |
| [13] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121):819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [14] |
Huang J, Wang Y, Zhao J. CRISPR editing in biological and biomedical investigation[J]. J Cell Physiol, 2018, 233(5):3875-3891.
doi: 10.1002/jcp.v233.5 URL |
| [15] |
Mashiko D, Young SA, Muto M, et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes[J]. Dev Growth Differ, 2014, 56(1):122-129.
doi: 10.1111/dgd.2014.56.issue-1 URL |
| [16] |
许祥, 董维鹏, 张少华, 等. 围脂滴蛋白基因CRISPR/Cas9载体的活性分析[J]. 生物技术通报, 2019, 35(11):89-95.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0357 |
| Xu X, Dong WP, Zhang SH, et al. Construction and activity analysis of the PLIN1 gene CRISPR/Cas9 vector[J]. Biotechnol Bull, 2019, 35(11):89-95. | |
| [17] | Morales PE, Bucarey JL, Espinosa A. Muscle lipid metabolism:role of lipid droplets and perilipins[J]. J Diabetes Res, 2017, 2017:1789395. |
| [18] | Sztalryd C, Brasaemle DL. The perilipin family of lipid droplet proteins:Gatekeepers of intracellular lipolysis[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2017, 1862(10 pt b):1221-1232. |
| [19] |
Maurizi G, Petäistö T, Maurizi A, et al. Key-genes regulating the liposecretion process of mature adipocytes[J]. J Cell Physiol, 2018, 233(5):3784-3793.
doi: 10.1002/jcp.26188 pmid: 28926092 |
| [20] | 赵志武, 王君实, 马敏, 等. 下调perilipin 1基因表达对3T3-L1细胞脂解的影响[J]. 中国生物工程杂志, 2016, 36(3):17-22. |
| Zhao ZW, Wang JS, Ma M, et al. Effect of down-regulated perilipin 1 gene expression on lipolysis of 3T3-L1 adipocytes[J]. China Biotechnol, 2016, 36(3):17-22. | |
| [21] | 张少华, 赵志武, 王君实, 等. 沉默PLIN1基因与异丙肾上腺素对3T3-L1脂肪细胞脂解的机制探究[J]. 现代预防医学, 2018, 45(11):2023-2027, 2038. |
| Zhang SH, Zhao ZW, Wang JS, et al. Combined effect of PLIN1 gene silencing and isoproterenol on lipolysis of 3T3-L1 adipocytes[J]. Mod Prev Med, 2018, 45(11):2023-2027, 2038. | |
| [22] |
Tansey JT, Sztalryd C, Gruia-Gray J, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity[J]. PNAS, 2001, 98(11):6494-6499.
doi: 10.1073/pnas.101042998 pmid: 11371650 |
| [23] |
Miyoshi H, Souza SC, Endo M, et al. Perilipin overexpression in mice protects against diet-induced obesity[J]. J Lipid Res, 2010, 51(5):975-982.
doi: 10.1194/jlr.M002352 URL |
| [1] | 史亚楠, 王德培, 王一川, 周昊, 薛鲜丽. 敲除msn2对米曲霉生长和发酵产曲酸的影响[J]. 生物技术通报, 2022, 38(8): 188-197. |
| [2] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [3] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| [4] | 王海杰, 王成稷, 郭洋, 王云, 陈艳娟, 梁敏, 王珏, 龚慧, 沈如凌. 基于CRSIPR/Cas9技术构建凝血因子8基因敲除小鼠模型及表型验证[J]. 生物技术通报, 2022, 38(10): 273-280. |
| [5] | 孙靖雅, 马玉超. 假单胞菌Tw224抗砷基因簇的功能研究[J]. 生物技术通报, 2022, 38(1): 141-149. |
| [6] | 王睿, 韩烈保. CRISPR/Cas9介导的二穗短柄草bdfls2敲除突变体的获得[J]. 生物技术通报, 2022, 38(1): 70-76. |
| [7] | 蒋成辉, 曾巧英, 王萌, 潘阳阳, 刘旭明, 尚天甜. CRISPR/Cas9构建srtA基因敲除的金黄色葡萄球菌[J]. 生物技术通报, 2020, 36(9): 253-265. |
| [8] | 梅芬, 李瑞玮, 张娟娟, 左荣霞, 邹云莲, 沈涛, 撒亚莲. 构建基于CRISPR/Cas9技术敲除ALOX5的质粒[J]. 生物技术通报, 2020, 36(3): 110-114. |
| [9] | 王松, 李鹏程, 白朝辉, 许宏鑫, 应研敏, 张墨, 白义春. 利用CRISPR/Cas9技术构建大鼠L2细胞α-ENaC基因敲除稳定细胞株[J]. 生物技术通报, 2020, 36(3): 115-123. |
| [10] | 叶洲杰, 王心睿. CRISPR系统转化医学研究进展[J]. 生物技术通报, 2020, 36(11): 188-197. |
| [11] | 吴航, 李智强, 刘文德. 稻瘟病菌分泌蛋白MoCDIE2诱导植物细胞死亡的功能分析[J]. 生物技术通报, 2020, 36(1): 15-22. |
| [12] | 陈和锋, 朱晁谊, 李爽. 产瓦伦西亚烯酿酒酵母的表达载体适配及发酵碳氮源优化[J]. 生物技术通报, 2020, 36(1): 209-219. |
| [13] | 杨雷, 叶洲杰, 李兆龙, 沈阳坤, 傅雅娟. 利用电转的方法对T细胞TET2基因敲除并探讨TET2对T细胞增殖的影响[J]. 生物技术通报, 2020, 36(1): 229-237. |
| [14] | 赵雅童, 何光明, 瓮茹茹, 石爱琴, 路福平, 李玉. 甘露醇磷酸化酶基因的敲除对D-甘露醇合成的影响[J]. 生物技术通报, 2019, 35(5): 118-124. |
| [15] | 明鹏飞, 黄莹莹, 董妍丽, 聂星灿, 冯士彬, 王希春, 程建波, 李锦春, 吴金节, 李玉. LKB1-AMPKα-SIRT1信号通路在奶牛脂肪组织脂代谢中的调控作用[J]. 生物技术通报, 2019, 35(2): 176-181. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||