








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 106-116.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0959
• 作物品质遗传与改良专题(专题主编: 刘巧泉 教授) • 上一篇 下一篇
林科运( ), 段钰晶, 王高升, 孙念礼, 方玉洁(
), 段钰晶, 王高升, 孙念礼, 方玉洁( ), 王幼平
), 王幼平
收稿日期:2021-07-21
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
方玉洁,女,博士,副教授,研究方向:油菜遗传育种与分子生物学;E-mail: yjfang@yzu.edu.cn作者简介:林科运,男,硕士研究生,研究方向:油菜遗传育种与分子生物学;E-mail: lkylab407@163.com
基金资助:
LIN Ke-yun( ), DUAN Yu-jing, WANG Gao-sheng, SUN Nian-li, FANG Yu-jie(
), DUAN Yu-jing, WANG Gao-sheng, SUN Nian-li, FANG Yu-jie( ), WANG You-ping
), WANG You-ping
Received:2021-07-21
Published:2022-04-26
Online:2022-05-06
摘要:
NF-YA是组成NF-Y转录因子的3个亚基之一,在植物中参与生长发育、胁迫响应以及与微生物互作等重要生物过程。通过克隆BnNF-YA1,并对BnNF-YA1进行生物信息学分析和表达模式分析,分离克隆其编码序列,构建过表达载体,转化甘蓝型油菜,通过阳性鉴定获得BnNF-YA1的转基因阳性植株,并对过表达BnNF-YA1的转基因植株进行了逆境表型鉴定。结果表明,BnNF-YA1的开放读码框为858 bp,编码285个氨基酸;烟草瞬时表达分析发现BnNF-YA1蛋白定位于细胞核内;酵母生化分析表明BnNF-YA1全长蛋白没有转录激活活性;表达分析结果显示,BnNF-YA1受PEG处理诱导表达,且在幼叶、花苞和受精后20 d的角果中表达水平较高;过表达BnNF-YA1的甘蓝型油菜对PEG和MV处理的耐受性提高,表明BnNF-YA1可能参与调控甘蓝型油菜对渗透胁迫和氧化胁迫的响应。
林科运, 段钰晶, 王高升, 孙念礼, 方玉洁, 王幼平. 甘蓝型油菜BnNF-YA1的克隆和功能鉴定[J]. 生物技术通报, 2022, 38(4): 106-116.
LIN Ke-yun, DUAN Yu-jing, WANG Gao-sheng, SUN Nian-li, FANG Yu-jie, WANG You-ping. Cloning and Functional Identification of BnNF-YA1 in Brassica napus L.[J]. Biotechnology Bulletin, 2022, 38(4): 106-116.
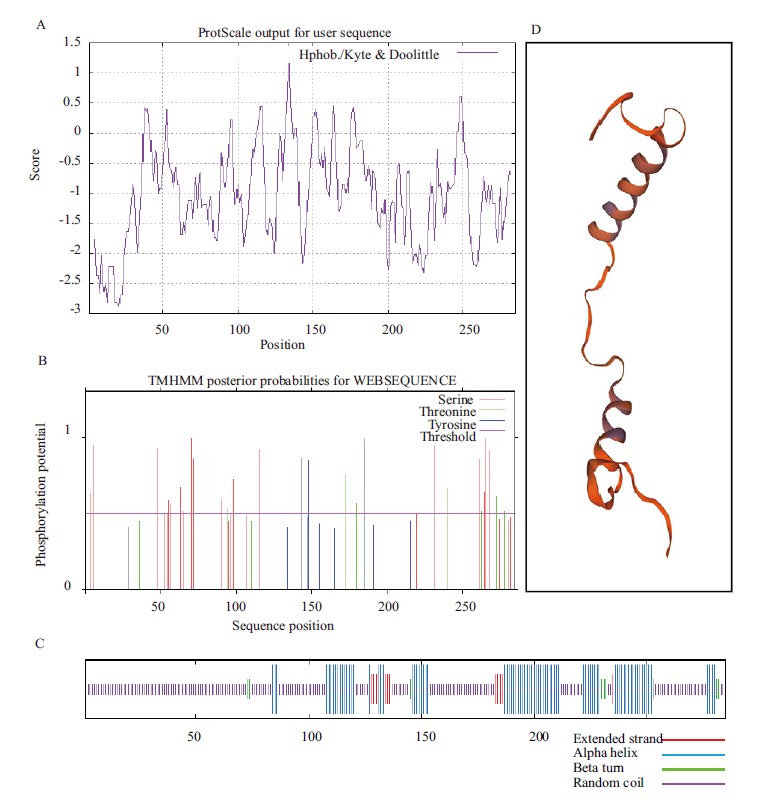
图1 BnNF-YA1亲/疏水性、磷酸化位点、二级结构、三级结构分析 A:BnNF-YA1亲/疏水性分布图;B:BnNF-YA1磷酸化位点预测;C:BnNF-YA1二级结构预测;D:BnNF-YA1三级结构预测
Fig. 1 Analysis of hydrophilicity/hydrophobicity,phos-phorylation site,secondary structure and tertiary structure of BnNF-YA1 A:Hydrophilic/hydrophobic analysis of BnNF-YA1. B:Prediction of phosphorylation sites of BnNF-YA1. C:Prediction of secondary structure of BnNF-YA1. D:Prediction of the tertiary structure of BnNF-YA1

图2 BnNF-YA1同源序列比对 Bn:油菜B. napus L;Bo:甘蓝B. oleracea;Br:白菜B. rapa;At:拟南芥Arabidopsis thaliana;Os:水稻Oryza sativa;Gm:大豆Glycine max;Zm:玉米Zea mays
Fig. 2 Multiple sequence alignments of BnNF-YA1 and its homologs
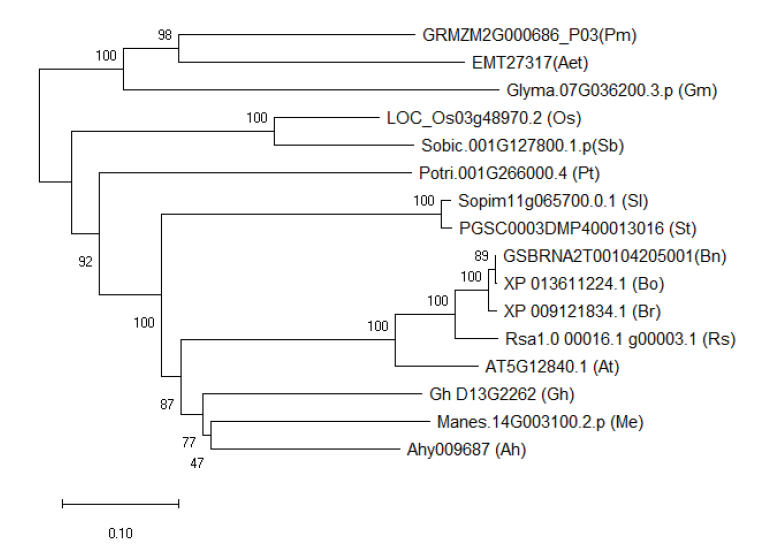
图3 NF-YA1系统发生分析 Zm:玉米Zea mays;Ata:山羊草Aegilops tauschii;Gm:大豆Glycine max;Os:水稻Oryza sativa;Sb:高粱Sorghum bicolor;Pt:白毛杨Populus tomentosa;Sl:番茄Solanum lycopersicum;St:马铃薯S. tuberosum;Bn:甘蓝型油菜Brassica napus L.;Bo:甘蓝B. oleracea;Br:白菜B. rapa;Rs:萝卜Raphanus sativus;At:拟南芥Arabidopsis thaliana;Gh:陆地棉Gossypium hirsutum;Me:木薯Manihot esculenta;Ah:花生Arachis hypogaea
Fig. 3 Phylogenetic analysis of NF-YA1 proteins
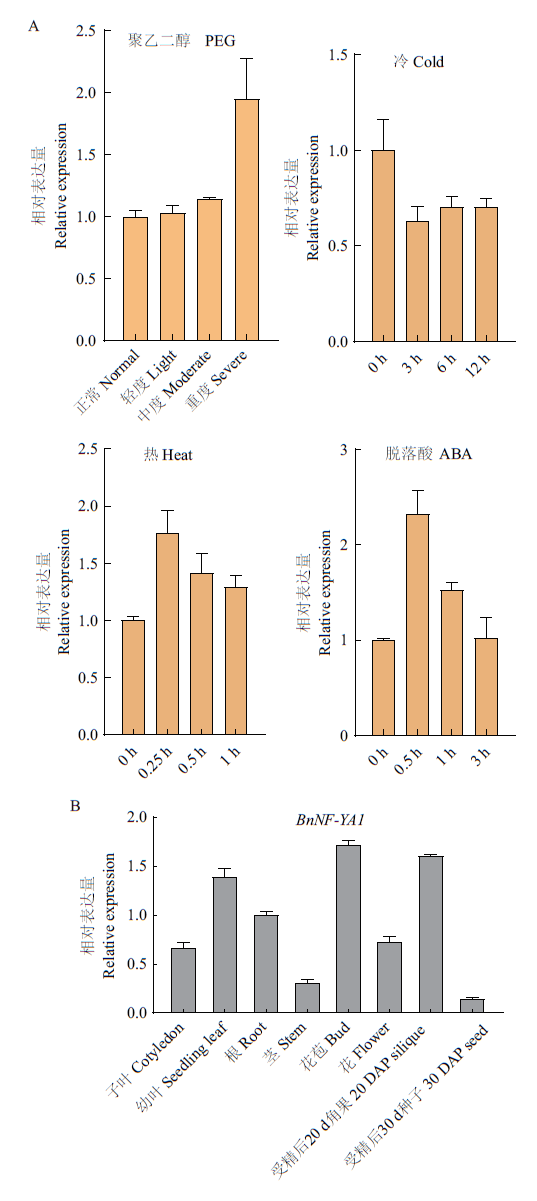
图4 BnNF-YA1表达模式分析 A:BnNF-YA1在聚乙二醇(PEG)、低温(cold)、高温(heat)以及脱落酸(ABA)处理条件下的表达变化情况;B:BnNF-YA1在甘蓝型油菜不同组织器官中的表达情况
Fig. 4 Expression profile of BnNF-YA1 A:Changes of BnNF-YA1 expression in B. napus under PEG,cold,heat,and ABA treatments. B:Spatial-temporal expression profile of BnNF-YA1 in the different organs of B. napus

图5 BnNF-YA1-OE过表达材料获得 A:过表达骨架载体示意图;B:BnNF-YA1 T0代过表达植株的PCR阳性鉴定;C:BnNF-YA1-OE转基因阳性植株RNA抽提检测;D:BnNF-YA1-OE转基因阳性植株反转录效果检测;E:BnNF-YA1-OE T0代过表达植株表达量鉴定
Fig. 5 Generation of BnNF-YA1-OE transgenic rapeseed plants A:Schematic diagram of the overexpresed vector backbone. B:PCR positive identification of overexpresed BnNF-YA1 in T0 generation. C:RNA detection of BnNF-YA1-OE transgenic positive plants. D:Reverse transcription detection of BnNF-YA1-OE transgenic positive plants. E:Expression of BnNF-YA1-OE in the T0 generation of overexpressed plants

图6 BnNF-YA1-OE T2代转基因植株子叶期表型鉴定 A:BnNF-YA1-OE T2代转基因植株子叶期表型;B:BnNF-YA1-OE T2代转基因植株下胚轴、根长及鲜重统计。误差表示标准差,*表示差异显著(P<0.05),**表示差异极显著(P<0.01)
Fig. 6 Phenotypic identification of T2 generation of BnNF-YA1-OE transgenic plants at the cotyledon stage A:Phenotype of T2 generation of BnNF-YA1-OE transgenic plants at the cotyledon stage. B:The hypocotyl length,root length and fresh weight of T2 generation of BnNF-YA1-OE transgenic plants. The error bars represent the standard deviation(SD). * represents significant difference(P<0.05),and ** represents extremely significant difference(P<0.01)
| [1] | 文雁成, 王汉中, 沈金雄, 等. 用SRAP标记分析中国甘蓝型油菜品种的遗传多样性和遗传基础[J]. 中国农业科学, 2006, 39(2):246-256. |
| Wen YC, Wang HZ, Shen JX, et al. Analysis of genetic diversity and genetic basis of Chinese rapeseed cultivars(Brassica napus L.)by sequence-related amplified polymorphism markers[J]. Sci Agric Sin, 2006, 39(2):246-256. | |
| [2] | 李利霞, 陈碧云, 闫贵欣, 等. 中国油菜种质资源研究利用策略与进展[J]. 植物遗传资源学报, 2020, 21(1):1-19. |
| Li LX, Chen BY, Yan GX, et al. Proposed strategies and current progress of research and utilization of oilseed rape germplasm in China[J]. J Plant Genet Resour, 2020, 21(1):1-19. | |
| [3] | 王汉中. 发展油菜生物柴油的潜力、问题与对策[J]. 中国油料作物学报, 2005, 27(2):74-76. |
| Wang HZ. Potential, problems and strategy of developing rapeseed biodiesel[J]. Chin J Oil Crop Scieves, 2005, 27(2):74-76. | |
| [4] | 唐湘如, 官春云. 几种酶活性与油菜油分和蛋白质及产量的关系[J]. 湖南农业大学学报, 2000, 26(1):37-40. |
| Tang XR, Guan CY. Relationship between activities of several enzymes and the oil, protein and yield of rapeseeds(Brassica napus)[J]. J Hunan Agric Univ, 2000, 26(1):37-40. | |
| [5] | 陈兆波, 余健. 我国油菜生产形势分析及科研对策研究[J]. 中国油料作物学报, 2010, 32(2):303-308. |
| Chen ZB, Yu J. Research strategies based on the analysis of rapeseed production in China[J]. Chin J Oil Crop Sci, 2010, 32(2):303-308. | |
| [6] | 王汉中. 我国油菜产业发展的历史回顾与展望[J]. 中国油料作物学报, 2010, 32(2):300-302. |
| Wang HZ. Review and future development of rapeseed industry in China[J]. Chin J Oil Crop Sci, 2010, 32(2):300-302. | |
| [7] |
Gyawali S, Parkin IAP, Steppuhn H, et al. Seedling, early vegetative, and adult plant growth of oilseed rapes(Brassica napus L.)under saline stress[J]. Can J Plant Sci, 2019, 99(6):927-941.
doi: 10.1139/cjps-2019-0023 URL |
| [8] |
Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress[J]. Plant Cell, 2002, 14(Suppl):S165-S183.
doi: 10.1105/tpc.000596 URL |
| [9] |
Zhu JK. Salt and drought stress signal transduction in plants[J]. Annu Rev Plant Biol, 2002, 53:247-273.
doi: 10.1146/annurev.arplant.53.091401.143329 URL |
| [10] |
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana[J]. Nature, 2000, 408(6814):796-815.
doi: 10.1038/35048692 URL |
| [11] | Singh D, Laxmi A. Transcriptional regulation of drought response:a tortuous network of transcriptional factors[J]. Front Plant Sci, 2015, 6:895. |
| [12] | Calvenzani V, Testoni B, Gusmaroli G, et al. Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits[J]. PLoS One, 2012, 7(8):e42902. |
| [13] |
Gnesutta N, Chiara M, Bernardini A, et al. The plant NF-Y DNA matrix in vitro and in vivo[J]. Plants, 2019, 8(10):406.
doi: 10.3390/plants8100406 URL |
| [14] |
Petroni K, Kumimoto RW, Gnesutta N, et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors[J]. Plant Cell, 2012, 24(12):4777-4792.
doi: 10.1105/tpc.112.105734 URL |
| [15] |
Mantovani R, Li XY, Pessara U, et al. Dominant negative analogs of NF-YA[J]. J Biol Chem, 1994, 269(32):20340-20346.
pmid: 8051128 |
| [16] |
Gnesutta N, Kumimoto RW, Swain S, et al. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer[J]. Plant Cell, 2017, 29(6):1516-1532.
doi: 10.1105/tpc.16.00864 URL |
| [17] |
Nardone V, Chaves-Sanjuan A, Nardini M. Structural determinants for NF-Y/DNA interaction at the CCAAT box[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(5):571-580.
doi: 10.1016/j.bbagrm.2016.09.006 URL |
| [18] |
Chaves-Sanjuan A, Gnesutta N, Gobbini A, et al. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants[J]. Plant J, 2021, 105(1):49-61.
doi: 10.1111/tpj.15038 URL |
| [19] |
Gnesutta N, Mantovani R, Fornara F. Plant flowering:imposing DNA specificity on histone-fold subunits[J]. Trends Plant Sci, 2018, 23(4):293-301.
doi: S1360-1385(17)30281-9 pmid: 29331540 |
| [20] |
Nardini M, Gnesutta N, Donati G, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination[J]. Cell, 2013, 152(1/2):132-143.
doi: 10.1016/j.cell.2012.11.047 URL |
| [21] |
Mach J. CONSTANS companion:CO binds the NF-YB/NF-YC dimer and confers sequence-specific DNA binding[J]. Plant Cell, 2017, 29(6):1183.
doi: 10.1105/tpc.17.00465 URL |
| [22] |
Li WX, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance[J]. Plant Cell, 2008, 20(8):2238-2251.
doi: 10.1105/tpc.108.059444 URL |
| [23] |
Lee DK, Kim HI, Jang G, et al. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner[J]. Plant Sci, 2015, 241:199-210.
doi: 10.1016/j.plantsci.2015.10.006 URL |
| [24] |
Zhang Q, Zhang J, Wei H, et al. Genome-wide identification of NF-YA gene family in cotton and the positive role of GhNF-YA10 and GhNF-YA23 in salt tolerance[J]. Int J Biol Macromol, 2020, 165(pt b):2103-2115.
doi: 10.1016/j.ijbiomac.2020.10.064 pmid: 33080263 |
| [25] |
Ni Z, Hu Z, Jiang Q, et al. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress[J]. Plant Mol Biol, 2013, 82(1/2):113-129.
doi: 10.1007/s11103-013-0040-5 URL |
| [26] |
Ma XJ, Yu TF, Li XH, et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants[J]. BMC Plant Biol, 2020, 20(1):123.
doi: 10.1186/s12870-020-02337-z URL |
| [27] |
Ma X, et al. Wheat NF-YA10 functions independently in salinity and drought stress[J]. Bioengineered, 2015, 6(4):245-247.
doi: 10.1080/21655979.2015.1054085 URL |
| [28] |
Lian C, Li Q, Yao K, et al. Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis[J]. Front Plant Sci, 2018, 9:954.
doi: 10.3389/fpls.2018.00954 URL |
| [29] | Pereira SLS, Martins CPS, Sousa AO, et al. Genome-wide characterization and expression analysis of Citrus NUCLEAR FACTOR-Y(NF-Y)transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance[J]. PLoS One, 2018, 13(6):e0199187. |
| [30] |
Sorin C, Declerck M, Christ A, et al. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis[J]. New Phytol, 2014, 202(4):1197-1211.
doi: 10.1111/nph.12735 URL |
| [31] |
Li C, Distelfeld A, Comis A, et al. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes[J]. Plant J, 2011, 67(5):763-773.
doi: 10.1111/j.1365-313X.2011.04630.x URL |
| [32] |
Fornari M, Calvenzani V, Masiero S, et al. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis[J]. PLoS One, 2013, 8(11):e82043.
doi: 10.1371/journal.pone.0082043 URL |
| [33] |
E Z, Li T, Zhang H, et al. A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots[J]. J Exp Bot, 2018, 69(10):2495-2510.
doi: 10.1093/jxb/ery087 URL |
| [34] |
Liu R, Wu M, Liu HL, et al. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus[J]. Physiol Plant, 2021, 171(3):309-327.
doi: 10.1111/ppl.13084 URL |
| [35] |
Guo Y, Niu S, El-Kassaby YA, et al. Transcriptome-wide isolation and expression of NF-Y gene family in male cone development and hormonal treatment of Pinus tabuliformis[J]. Physiol Plant, 2021, 171(1):34-47.
doi: 10.1111/ppl.13183 URL |
| [36] |
Mai YT, Shui LY, Huo KS, et al. Genome-wide characterization of the NUCLEAR FACTOR-Y(NF-Y)family in Citrus grandis identified CgNF-YB9 involved in the fructose and glucose accumulation[J]. Genes Genom, 2019, 41(11):1341-1355.
doi: 10.1007/s13258-019-00862-2 URL |
| [37] | Maheshwari P, Kummari D, Palakolanu SR, et al. Genome-wide identification and expression profile analysis of nuclear factor Y family genes in Sorghum bicolor L. (Moench)[J]. PLoS One, 2019, 14(9):e0222203. |
| [38] |
Liang M, Yin X, Lin Z, et al. Identification and characterization of NF-Y transcription factor families in Canola(Brassica napus L.)[J]. Planta, 2014, 239(1):107-126.
doi: 10.1007/s00425-013-1964-3 URL |
| [39] | Xu L, Lin Z, Tao Q, et al. Multiple nuclear factor Y transcription factors respond to abiotic stress in Brassica napus L.[J]. PLoS One, 2014, 9(10):e111354. |
| [40] |
Kumar S, Stecher G, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [41] | 石淑稳, 周永明, 孙学成, 等. 甘蓝型油菜遗传转化体系的研究[J]. 华中农业大学学报, 1998, 17(3):205-210. |
| Shi SW, Zhou YM, Sun XC, et al. Transformation of Brassica napus with herbicide resistance gene[J]. J Huazhong Agric, 1998, 17 (3):205-210. | |
| [42] | 李佳, 沈斌章, 韩继祥, 等. 一种有效提取油菜叶片总DNA的方法[J]. 华中农业大学学报, 1994, 13(5):521-523. |
| Li J, Shen BZ, Han JX, et al. A effective procedure for extracting total DNA in rape[J]. J Huazhong:Central China Agric Univ, 1994, 13(5):521-523. | |
| [43] | Rao XY, et al. An improvement of the 2(-delta delta CT)method for quantitative real-time polymerase chain reaction data analysis[J]. Biostat Bioinforma Biomath, 2013, 3(3):71-85. |
| [44] | 王寻, 冯资权, 等. 苹果NF-YA转录因子家族的生物信息学和表达分析[J]. 植物生理学报, 2021, 57(1):69-84. |
|
Wang X, Feng ZQ, You CX, et al. Bioinformatics and expression analysis of NF-YA transcription factor family in apple[J]. Plant Physiol J, 2021, 57(1):69-84.
doi: 10.1104/pp.57.1.69 URL |
|
| [45] | 任旭洋. 四个滨海耐盐油菜转录因子(BnNF-YA2/3/9/12)的耐逆功能分析[D]. 南京:南京农业大学, 2017. |
| Ren XY. Analysis of stress tolerance function of four transcription factors(BnNF-YA2/3/9/12)in coastal salt tolerant rapeseed[D]. Nanjing:Nanjing Agricultural University, 2017. | |
| [46] |
黄锁, 胡利芹, 徐东北, 等. 谷子转录因子SiNF-YA5通过ABA非依赖途径提高转基因拟南芥耐盐性[J]. 作物学报, 2016, 42(12):1787-1797.
doi: 10.3724/SP.J.1006.2016.01787 |
|
Huang S, Hu LQ, Xu DB, et al. Transcription factor SiNF-YA5 from foxtail millet(Setaria italica)conferred tolerance to high-salt stress through ABA-independent pathway in transgenic Arabidopsis[J]. Acta Agron Sin, 2016, 42(12):1787-1797.
doi: 10.3724/SP.J.1006.2016.01787 URL |
|
| [47] | 王蓓. 菊花DgbZIP2和DgNF-YA1基因的克隆与转基因菊花耐寒性研究[D]. 雅安:四川农业大学, 2019. |
| Wang B. Cloning and functional identification of DgbZIP2 and DgNF-YA1 in Chrysanthemum[D]. Yan’an:Sichuan Agricultural University, 2019. |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 刘保财, 陈菁瑛, 张武君, 黄颖桢, 赵云青, 刘剑超, 危智诚. 多花黄精种子微根茎基因表达特征分析[J]. 生物技术通报, 2023, 39(8): 220-233. |
| [6] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [7] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [8] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [9] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [10] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [11] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [12] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [13] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [14] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [15] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||