








生物技术通报 ›› 2022, Vol. 38 ›› Issue (7): 51-61.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1524
陈佳敏1,2( ), 刘永杰2, 马锦绣2, 李丹2, 公杰2, 赵昌平2, 耿洪伟1(
), 刘永杰2, 马锦绣2, 李丹2, 公杰2, 赵昌平2, 耿洪伟1( ), 高世庆2(
), 高世庆2( )
)
收稿日期:2021-12-08
出版日期:2022-07-26
发布日期:2022-08-09
作者简介:陈佳敏,女,硕士研究生,研究方向:农艺与种业;E-mail:基金资助:
CHEN Jia-min1,2( ), LIU Yong-jie2, MA Jin-xiu2, LI Dan2, GONG Jie2, ZHAO Chang-ping2, GENG Hong-wei1(
), LIU Yong-jie2, MA Jin-xiu2, LI Dan2, GONG Jie2, ZHAO Chang-ping2, GENG Hong-wei1( ), GAO Shi-qing2(
), GAO Shi-qing2( )
)
Received:2021-12-08
Published:2022-07-26
Online:2022-08-09
摘要:
旨在筛选参与小麦杂交种干旱胁迫的组蛋白甲基化酶基因,为二系杂交小麦抗逆性改良提供功能标记。通过网站公开数据库和比较基因组学策略对小麦TaHMT家族175个基因进行筛选获得8个受干旱胁迫诱导表达候选基因(TaHMT21、TaHMT24、TaHMT31、TaHMT42、TaHMT49、TaHMT105、TaHMT143、TaHMT157)。采用生物信息学方法对8个TaHMTs理化性质、蛋白结构、系统发育关系、基因结构和保守基序等进行了系统分析。进一步利用干旱处理的小麦杂交组合(BS278*09YH91-5及父母本)为试材,对8个组蛋白甲基化酶候选基因进行荧光定量PCR验证,发现它们受不同程度干旱胁迫诱导表达,且随着干旱胁迫时间延长基因表达水平呈先上调后下调的模式,其中TaHMT21、TaHMT24、TaHMT42基因表达与杂交组合的抗旱表型相符合,从而推测这些基因可能参与调控杂交种的抗旱性。上述结果为深入研究小麦TaHMT家族基因在杂交种抗旱性中发挥的功能提供了重要线索,同时也为小麦分子育种提供优异抗逆基因资源。
陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61.
CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat[J]. Biotechnology Bulletin, 2022, 38(7): 51-61.
| 引物名称Primer name | 上游引物序列 Forward primer sequence(5'-3') | 下游引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| actin | GTTGGTGATGAGGCCCAATC | GTGCTACACGGAGCTCATTG |
| TaHMT21 | TGCCGCTGTGGTGTATACTG | CAGCCAAAAGACCCCATCCA |
| TaHMT24 | TAAGGCTAGGGGCAACTCCT | CCATCCAACTCAGCAGTCGT |
| TaHMT31 | GTTTGGTGGTGTGATGGTGC | TGCTGGAATGGTGTCCTTCG |
| TaHMT42 | GGTCTGCGCAAGCAATTGAA | TCGTACAGGGCGTCGATTTC |
| TaHMT49 | TTCTTCACGAGCAGGAAGGT | TTCCTCGGCAGTACTTGCTT |
| TaHMT105 | AGACCTTAGAGAGCGGCGTA | ATGCTCATCCCCGAGAACAC |
| TaHMT143 | TGATTGCAGCATGGACCAGT | GCAAACAGCCCCAAAGTACG |
| TaHMT157 | CTACGGTTGCATCAAGCTCA | GCATCTCACAGTCCTCGTCA |
表1 实时定量PCR引物序列
Table 1 Primer sequences for real-time quantitative PCR
| 引物名称Primer name | 上游引物序列 Forward primer sequence(5'-3') | 下游引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| actin | GTTGGTGATGAGGCCCAATC | GTGCTACACGGAGCTCATTG |
| TaHMT21 | TGCCGCTGTGGTGTATACTG | CAGCCAAAAGACCCCATCCA |
| TaHMT24 | TAAGGCTAGGGGCAACTCCT | CCATCCAACTCAGCAGTCGT |
| TaHMT31 | GTTTGGTGGTGTGATGGTGC | TGCTGGAATGGTGTCCTTCG |
| TaHMT42 | GGTCTGCGCAAGCAATTGAA | TCGTACAGGGCGTCGATTTC |
| TaHMT49 | TTCTTCACGAGCAGGAAGGT | TTCCTCGGCAGTACTTGCTT |
| TaHMT105 | AGACCTTAGAGAGCGGCGTA | ATGCTCATCCCCGAGAACAC |
| TaHMT143 | TGATTGCAGCATGGACCAGT | GCAAACAGCCCCAAAGTACG |
| TaHMT157 | CTACGGTTGCATCAAGCTCA | GCATCTCACAGTCCTCGTCA |

图1 小麦TaHMT候选基因干旱胁迫表达热图 A:在Wheat Expression Brower数据库和WheatOmics数据库上差异表达基因与同源基因的韦恩图;B:8个候选基因在Wheat Expression Brower数据库的表达热图;C:8个候选基因在WheatOmics数据库的表达热图。红色字体为候选基因
Fig.1 Heatmap of TaHMT candidate gene expressions under drought stress in wheat A:Venn diagram of differentially expressed genes and homologous genes on Wheat Expression Brower database and WheatOmics database. B:Expression heatmap of 8 candidate genes in Wheat Expression Brower database. C:Expression heatmap of 8 candidate genes in WheatOmics database. The red font is the candidate gene
| 基因名称Gene name | TaHMT21 | TaHMT24 | TaHMT31 | TaHMT42 | TaHMT49 | TaHMT105 | TaHMT143 | TaHMT157 |
|---|---|---|---|---|---|---|---|---|
| 基因ID号Gene ID | TraesCS2A02G302100 | TraesCS2A02G457100 | TraesCS2B02G317900 | TraesCS2D02G300800 | TraesCS2D02G566400 | TraesCS5B02G163000 | TraesCS7B02G262900 | TraesCS2A02G147100 |
| 染色体位置Chromosome location | 2A:517 803 993-517 808 473(+) | 2A:705 740 079-705 742 875(+) | 2B:453 747 757-453 752 627(+) | 2D:383 285 805-383 290 377(+) | 2D:635 593 374-635 597 307(-) | 5B:300 380 740-300 397 790(-) | 7B:483 766 747-483 769 174(-) | 2A:93 926 568-93 930 236(-) |
| 亚细胞定位Subcellular localization | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| 氨基酸数目Number of amino acids/aa | 502 | 476 | 529 | 502 | 737 | 1086 | 390 | 601 |
| 分子量Molecular weight/kD | 56 612.99 | 53 938.51 | 59 323.00 | 56 560.96 | 80 411.70 | 122 007.19 | 42 293.23 | 65 280.55 |
| 理论等电点Theoretical pI | 4.77 | 5.47 | 4.78 | 4.80 | 7.09 | 7.59 | 4.44 | 4.48 |
| 负电荷残基数(Asp+Glu) Number of negatively charged residues(Asp+Glu) | 83 | 64 | 84 | 82 | 84 | 146 | 65 | 99 |
| 正电荷残基数(Arg+Lys) Number of positively charged residues(Arg+Lys) | 53 | 53 | 53 | 53 | 83 | 148 | 30 | 50 |
| 总原子数Total number of atoms | 7 801 | 7 540 | 8 162 | 7 794 | 11 137 | ------ | 5 789 | 8 992 |
| 分子式Formula | C2458H3830N684O 805S24 | C2392H3751N657O 716S24 | C2572H4003N717O 843S27 | C2458H3826N684O 802S24 | C3478H5503N1035O 1080S41 | ------ | C1833H2830N506O 594S26 | C2859H4408N762O 937S26 |
| 不稳定指数Instability index | 51.39 | 43.26 | 49.28 | 53.18 | 49.44 | 54.44 | 54.59 | 38.91 |
| 脂溶指数Aliphatic index | 71.14 | 90.82 | 71.02 | 71.53 | 68.41 | 76.19 | 75.95 | 75.61 |
| 亲水性GRAVY | -0.631 | -0.219 | -0.584 | -0.608 | -0.511 | -0.514 | -0.301 | -0.285 |
表2 TaHMT干旱胁迫候选基因基本信息
Table 2 Basic information of TaHMT drought stress candidate genes
| 基因名称Gene name | TaHMT21 | TaHMT24 | TaHMT31 | TaHMT42 | TaHMT49 | TaHMT105 | TaHMT143 | TaHMT157 |
|---|---|---|---|---|---|---|---|---|
| 基因ID号Gene ID | TraesCS2A02G302100 | TraesCS2A02G457100 | TraesCS2B02G317900 | TraesCS2D02G300800 | TraesCS2D02G566400 | TraesCS5B02G163000 | TraesCS7B02G262900 | TraesCS2A02G147100 |
| 染色体位置Chromosome location | 2A:517 803 993-517 808 473(+) | 2A:705 740 079-705 742 875(+) | 2B:453 747 757-453 752 627(+) | 2D:383 285 805-383 290 377(+) | 2D:635 593 374-635 597 307(-) | 5B:300 380 740-300 397 790(-) | 7B:483 766 747-483 769 174(-) | 2A:93 926 568-93 930 236(-) |
| 亚细胞定位Subcellular localization | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| 氨基酸数目Number of amino acids/aa | 502 | 476 | 529 | 502 | 737 | 1086 | 390 | 601 |
| 分子量Molecular weight/kD | 56 612.99 | 53 938.51 | 59 323.00 | 56 560.96 | 80 411.70 | 122 007.19 | 42 293.23 | 65 280.55 |
| 理论等电点Theoretical pI | 4.77 | 5.47 | 4.78 | 4.80 | 7.09 | 7.59 | 4.44 | 4.48 |
| 负电荷残基数(Asp+Glu) Number of negatively charged residues(Asp+Glu) | 83 | 64 | 84 | 82 | 84 | 146 | 65 | 99 |
| 正电荷残基数(Arg+Lys) Number of positively charged residues(Arg+Lys) | 53 | 53 | 53 | 53 | 83 | 148 | 30 | 50 |
| 总原子数Total number of atoms | 7 801 | 7 540 | 8 162 | 7 794 | 11 137 | ------ | 5 789 | 8 992 |
| 分子式Formula | C2458H3830N684O 805S24 | C2392H3751N657O 716S24 | C2572H4003N717O 843S27 | C2458H3826N684O 802S24 | C3478H5503N1035O 1080S41 | ------ | C1833H2830N506O 594S26 | C2859H4408N762O 937S26 |
| 不稳定指数Instability index | 51.39 | 43.26 | 49.28 | 53.18 | 49.44 | 54.44 | 54.59 | 38.91 |
| 脂溶指数Aliphatic index | 71.14 | 90.82 | 71.02 | 71.53 | 68.41 | 76.19 | 75.95 | 75.61 |
| 亲水性GRAVY | -0.631 | -0.219 | -0.584 | -0.608 | -0.511 | -0.514 | -0.301 | -0.285 |
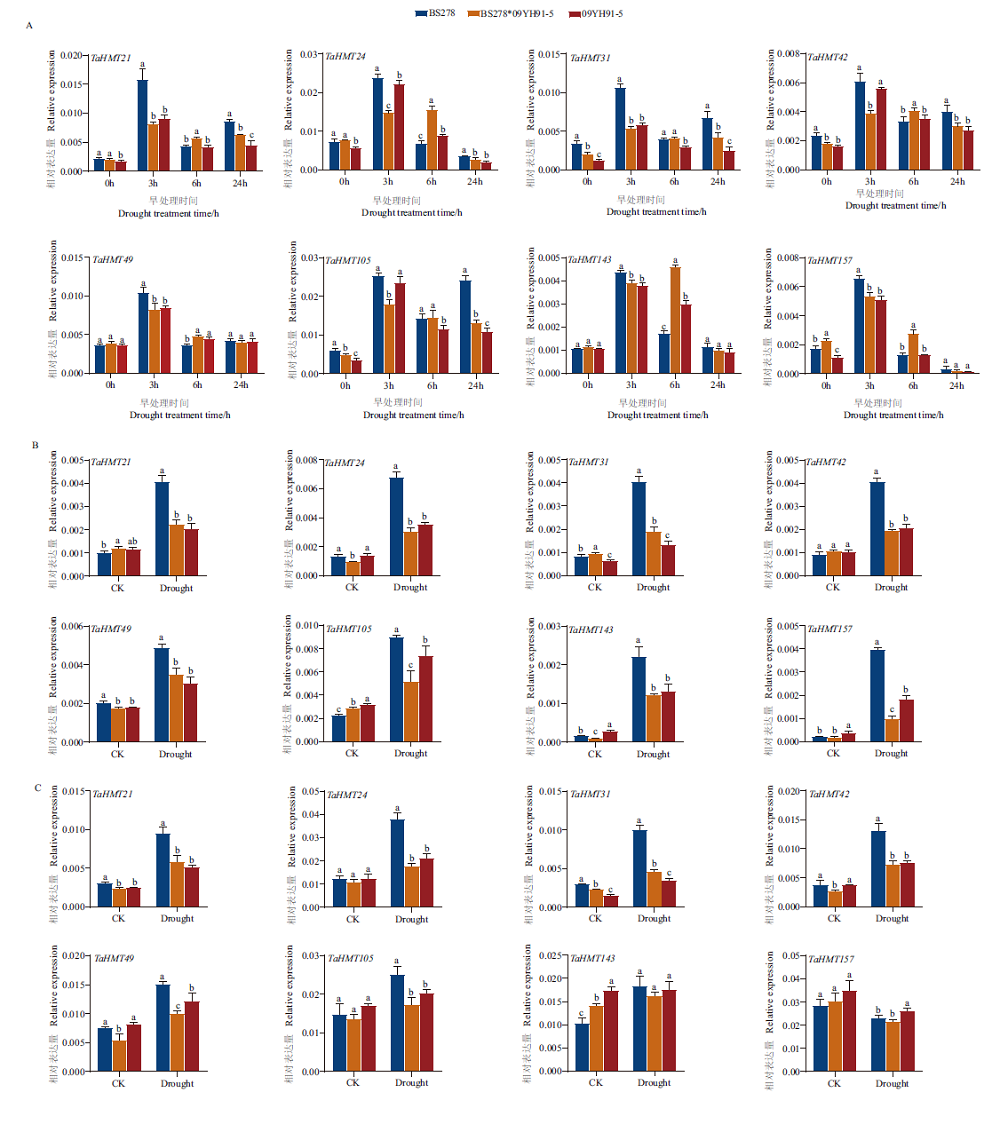
图6 TaHMT干旱胁迫下的表达模式分析 A:营养生长期小麦叶片中基因的表达;B:苗期小麦地上部中基因的表达;C:苗期小麦地下部中基因的表达。图中的误差线表示标准偏差。小写字母表示同一时间不同材料之间的显著差异(P<0.05)
Fig.6 Expressions of TaHMT under drought stress A:Gene expression in the wheat leaves during vegetative growth. B:Gene expression in the shoot of wheat at seedling stage. C:Gene expression in the underground part of wheat at seedling stage. The error line in the figure refers to the standard deviation. The lowercase letters indicate significant differences between different materials at the same time(P<0.05)
| [1] | Goldberg AD, Allis CD, Bernstein E. Epigenetics:a landscape takes shape[J]. Cell, 2007, 128(4):635-638. |
| [2] | Huang H, Sabari BR, Garcia BA, et al. SnapShot:histone modifications[J]. Cell, 2014, 159(2):458-458. e1. |
| [3] | Lawrence M, Daujat S, Schneider R. Lateral thinking:how histone modifications regulate gene expression[J]. Trends Genet, 2016, 32(1):42-56. |
| [4] | Jenuwein T, Allis CD. Translating the histone code[J]. Science, 2001, 293(5532):1074-1080. |
| [5] | Strahl BD, Allis CD. The language of covalent histone modifications[J]. Nature, 2000, 403(6765):41-45. |
| [6] | Feng Q, Wang HB, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain[J]. Curr Biol, 2002, 12(12):1052-1058. |
| [7] | Yeates TO. Structures of SET domain proteins:protein lysine methyltransferases make their mark[J]. Cell, 2002, 111(1):5-7. |
| [8] | Ng DWK, Wang T, Chandrasekharan MB, et al. Plant SET domain-containing proteins:structure, function and regulation[J]. Biochim Biophys Acta, 2007, 1769(5/6):316-329. |
| [9] | Ahmad A, Dong YZ, Cao XF. Characterization of the PRMT gene family in rice reveals conservation of arginine methylation[J]. PLoS One, 2011, 6(8):e22664. |
| [10] | Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation[J]. Prog Nucleic Acid Res Mol Biol, 1998, 61:65-131. |
| [11] | Boisvert FM, Chénard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation[J]. Sci STKE, 2005, 2005(271):re2. |
| [12] | Bedford MT, Clarke SG. Protein arginine methylation in mammals:who, what, and why[J]. Mol Cell, 2009, 33(1):1-13. |
| [13] | Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants[J]. Curr Opin Plant Biol, 2009, 12(2):133-139. |
| [14] | Luo M, Liu XC, Singh P, et al. Chromatin modifications and remodeling in plant abiotic stress responses[J]. Biochim Biophys Acta, 2012, 1819(2):129-136. |
| [15] | Kumar SV, Wigge PA. H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis[J]. Cell, 2010, 140(1):136-147. |
| [16] | Ding Y, Lapko H, Ndamukong I, et al. The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought[J]. Plant Signal Behav, 2009, 4(11):1049-1058. |
| [17] | Chen K, Du KX, Shi YC, et al. H3K36 methyltransferase SDG708 enhances drought tolerance by promoting abscisic acid biosynthesis in rice[J]. New Phytol, 2021, 230(5):1967-1984. |
| [18] | Sun XW, Chen L, et al. Histone methyltransferase SDG8 in dehydr-ation stress[J]. J Univ Sci Technol China, 2021, 51(2):140-146. |
| [19] | 赵燕昊, 曹跃芬, 孙威怡, 等. 小麦抗旱研究进展[J]. 植物生理学报, 2016, 52(12):1795-1803. |
| Zhao YH, Cao YF, Sun WY, et al. The research advances in drought resistance in wheat[J]. Plant Physiol J, 2016, 52(12):1795-1803. | |
| [20] | 杨梅. 普通小麦转TaEBP基因系抗旱特性研究[D]. 杨凌: 西北农林科技大学, 2012. |
| Yang M. Study on drought-resistance of transgenic wheat with TaEBP gene[D]. Yangling: Northwest A & F University, 2012. | |
| [21] | Edae EA, Byrne PF, et al. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes[J]. Theor Appl Genet, 2014, 127(4):791-807. |
| [22] | Chen CJ, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8):1194-1202. |
| [23] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]// Walker JM. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005:571-607. |
| [24] | Chou KC, Shen HB. Cell-PLoc:a package of Web servers for predicting subcellular localization of proteins in various organisms[J]. Nat Protoc, 2008, 3(2):153-162. |
| [25] | Geourjon C, Deléage G. SOPMA:significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Comput Appl Biosci, 1995, 11(6):681-684. |
| [26] | Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis[J]. Nat Protoc, 2015, 10(6):845-858. |
| [27] | Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers[J]. Proc Int Conf Intell Syst Mol Biol, 1994, 2:28-36. |
| [28] | Kumar S, Stecher G, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6):1547-1549. |
| [29] | He ZL, Zhang HK, Gao SH, et al. Evolview v2:an online visualization and management tool for customized and annotated phylogenetic trees[J]. Nucleic Acids Res, 2016, 44(W1):W236-W241. |
| [30] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1):325-327. |
| [31] | Ding Y, Avramova Z, Fromm M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways[J]. Plant J, 2011, 66(5):735-744. |
| [32] | Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat[J]. Front Plant Sci, 2014, 5:170. |
| [33] | Cao XY, Costa LM, Biderre-Petit C, et al. Abscisic acid and stress signals induce viviparous1 expression in seed and vegetative tissues of maize[J]. Plant Physiol, 2007, 143(2):720-731. |
| [34] | Yang WT, Baek D, Yun DJ, et al. Rice OsMYB5P improves plant phosphate acquisition by regulation of phosphate transporter[J]. PLoS One, 2018, 13(3):e0194628. |
| [35] | Wu JD, Jiang YL, Liang YN, et al. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants[J]. Plant Physiol Biochem, 2019, 137:179-188. |
| [36] | Xu ZJ, Sun ML, Jiang XF, et al. Glycinebetaine biosynthesis in response to osmotic stress depends on jasmonate signaling in watermelon suspension cells[J]. Front Plant Sci, 2018, 9:1469. |
| [37] | 马超, 张均, 等. 外源MeJA对花后干旱胁迫下小麦光合特性的影响[J]. 麦类作物学报, 2018, 38(5):563-571. |
| Ma C, Zhang J, et al. Effect of exogenous methyl jasmonate on photosynthetic characteristics in wheat under drought stress after anthesis[J]. J Triticeae Crops, 2018, 38(5):563-571. | |
| [38] | Rouster J, Leah R, et al. Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain[J]. Plant J, 1997, 11(3):513-523. |
| [39] | Aquea F, Vega A, Timmermann T, et al. Genome-wide analysis of the set domain group family in grapevine[J]. Plant Cell Rep, 2011, 30(6):1087-1097. |
| [40] | Napsucialy-Mendivil S, Alvarez-Venegas R, Shishkova S, et al. Arabidopsis homolog of trithorax1(ATX1)is required for cell production, patterning, and morphogenesis in root development[J]. J Exp Bot, 2014, 65(22):6373-6384. |
| [41] | Choi SC, Lee S, Kim SR, et al. Trithorax group protein Oryza sativa trithorax1 controls flowering time in rice via interaction with early heading date3[J]. Plant Physiol, 2014, 164(3):1326-1337. |
| [42] | Hang RL, Liu CY, Ahmad A, et al. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing[J]. PNAS, 2014, 111(45):16190-16195. |
| [43] | Han YF, et al. SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis[J]. Cell Res, 2014, 24(12):1445-1465. |
| [44] | Luo YX, Han YF, Zhao QY, et al. Sumoylation of SUVR2 contributes to its role in transcriptional gene silencing[J]. Sci China Life Sci, 2018, 61(2):235-243. |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [4] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [5] | 丁凯鑫, 王立春, 田国奎, 王海艳, 李凤云, 潘阳, 庞泽, 单莹. 烯效唑缓解植物干旱损伤的研究进展[J]. 生物技术通报, 2023, 39(6): 1-11. |
| [6] | 王春语, 李政君, 王平, 张丽霞. 高粱表皮蜡质缺失突变体sb1抗旱生理生化分析[J]. 生物技术通报, 2023, 39(5): 160-167. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [9] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [10] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [11] | 于波, 秦晓惠, 赵杨. 植物感应干旱信号的机制[J]. 生物技术通报, 2023, 39(11): 6-17. |
| [12] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [13] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [14] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [15] | 孔德真, 聂迎彬, 崔凤娟, 桑伟, 徐红军, 田笑明. 杂交小麦制种研究现状及展望[J]. 生物技术通报, 2023, 39(1): 95-103. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||