








生物技术通报 ›› 2023, Vol. 39 ›› Issue (11): 328-339.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0049
收稿日期:2023-01-18
出版日期:2023-11-26
发布日期:2023-12-20
通讯作者:
兰海燕,女,博士,教授,研究方向:植物抗逆分子生物学;E-mail: Lanhaiyan@xju.edu.cn作者简介:鄢梦雨,女,硕士研究生,研究方向:植物抗逆分子生物学;E-mail: 1610372465@qq.com
基金资助:
YAN Meng-yu( ), WEI Xiao-wei, CAO Jing, LAN Hai-yan(
), WEI Xiao-wei, CAO Jing, LAN Hai-yan( )
)
Received:2023-01-18
Published:2023-11-26
Online:2023-12-20
摘要:
碱性螺旋-环-螺旋(basic belix-loop-helix,bHLH)转录因子是真核生物中最大的转录因子家族之一,其成员广泛参与植物生长、发育和胁迫响应。前期在异子蓬(Suaeda aralocaspica)光合作用关键酶PEPC-1的研究中筛选到一个可能与SaPEPC-1启动子相互作用的bHLH转录因子SabHLH169,探明该基因功能将为后期异子蓬bHLH基因的深入研究奠定基础。通过克隆获得该基因,采用生物信息学、实时荧光定量PCR等方法,初步探究SabHLH169基因的耐旱功能。结果显示,SabHLH169包含2 100 bp核苷酸,编码699个氨基酸,其蛋白质C末端具有典型的bHLH结构域,原核表达和Western blot分析表明,SabHLH169重组蛋白能够正确表达,其分子量大小与预测结果一致。过表达SabHLH169的拟南芥转基因株系在萌发期具有更高的耐旱性;干旱胁迫下,转基因拟南芥中3个干旱胁迫相关基因AtRD22、AtRD29A和AtDREB2A以及3个光合作用关键酶基因AtLhcb2、AtRubicso和AtPEPC的表达水平显著升高,且AtPEPC酶活性被显著增强。由此推测,SabHLH169基因可能在干旱胁迫响应中具有潜在功能。
鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339.
YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress[J]. Biotechnology Bulletin, 2023, 39(11): 328-339.
| 名称 Name | 上游引物序列 Forward primer(5'-3') | 下游引物序列 Reverse primer(5'-3') |
|---|---|---|
| SabHLH169(PCR) | ATGGCTACTCACTTGCAGCAGTTGCTTC | TCAGGCATTAGTTTTTGATTGAAGTAGTTGG |
| pSuper1300-SabHLH169 | GCGTCGACATGGCTACTCACTTGCAGCAGTTGCT | CGGGATCCTCAGGCATTAGTTTTTGATTGAAGTA |
| pMal-c2X-SabHLH169 | CGGGATCCATGGCTACTCACTTGCAGCAGTTGCTTC | GCGTCGACTCAGGCATTAGTTTTTGATTGAAGTAGTT |
| AtHPT | GGTCGCGGAGGCTATGGATGC | GCTTCTGCGGGCGATTTGTGT |
| AtRD22(qRT-PCR) | GACTTTCGATTTTACCGACGAG | CGCTACCGGTTTTACCTTTATG |
| AtDREB2A(qRT-PCR) | CGTTTCAGGATGAGATGTGTGA | CTCATCGTGCATATAAAACGCA |
| AtRD29A(qRT-PCR) | TTCTGTAAGGACGACGTTTACA | CGTACTCGTTACATCCTCTGTT |
| AtACTIN(qRT-PCR) | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| AtPEPC(qRT-PCR) | AGCCTTCAGGGAACCACAAT | CTCCAAAGACGGGTCGCATG |
| AtRubisco(qRT-PCR) | TGGCTTCCTCTATGCTCTCTTC | ACACTTGAGCGGAGTCGGTGCA |
| AtLhcb2(qRT-PCR) | ATGTTGGGTGCTCTCGGATG | CGCGTGGATCAAGTTAGGGT |
| AtRCA(qRT-PCR) | GGCCGCCGCAGTTTCCACCG | AAAGTTGTAGACACAGGTTCCA |
表1 本文所用引物序列
Table 1 Primer sequences used in this study
| 名称 Name | 上游引物序列 Forward primer(5'-3') | 下游引物序列 Reverse primer(5'-3') |
|---|---|---|
| SabHLH169(PCR) | ATGGCTACTCACTTGCAGCAGTTGCTTC | TCAGGCATTAGTTTTTGATTGAAGTAGTTGG |
| pSuper1300-SabHLH169 | GCGTCGACATGGCTACTCACTTGCAGCAGTTGCT | CGGGATCCTCAGGCATTAGTTTTTGATTGAAGTA |
| pMal-c2X-SabHLH169 | CGGGATCCATGGCTACTCACTTGCAGCAGTTGCTTC | GCGTCGACTCAGGCATTAGTTTTTGATTGAAGTAGTT |
| AtHPT | GGTCGCGGAGGCTATGGATGC | GCTTCTGCGGGCGATTTGTGT |
| AtRD22(qRT-PCR) | GACTTTCGATTTTACCGACGAG | CGCTACCGGTTTTACCTTTATG |
| AtDREB2A(qRT-PCR) | CGTTTCAGGATGAGATGTGTGA | CTCATCGTGCATATAAAACGCA |
| AtRD29A(qRT-PCR) | TTCTGTAAGGACGACGTTTACA | CGTACTCGTTACATCCTCTGTT |
| AtACTIN(qRT-PCR) | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| AtPEPC(qRT-PCR) | AGCCTTCAGGGAACCACAAT | CTCCAAAGACGGGTCGCATG |
| AtRubisco(qRT-PCR) | TGGCTTCCTCTATGCTCTCTTC | ACACTTGAGCGGAGTCGGTGCA |
| AtLhcb2(qRT-PCR) | ATGTTGGGTGCTCTCGGATG | CGCGTGGATCAAGTTAGGGT |
| AtRCA(qRT-PCR) | GGCCGCCGCAGTTTCCACCG | AAAGTTGTAGACACAGGTTCCA |

图1 异子蓬总RNA的提取及SabHLH169 ORF的扩增 A:异子蓬幼苗总RNA.1, 2:两个总RNA样品;B:SabHLH169 ORF的PCR扩增. M1, M2:DL 2000, DL5000 DNA分子量标准
Fig. 1 Extraction of total RNA and PCR amplification of SabHLH169 ORF in S. aralocaspica A: Total RNA of S. aralocaspica seedlings. 1,2: Two samples of total RNA; B: PCR amplification of SabHLH169 ORF; M1, M2: DL2000, DL5000 DNA markers

图2 异子蓬SabHLH169氨基酸序列分析 N端下划线部分表示bHLH-MYC_N结构域;C端下划线部分表示HLH结构域
Fig. 2 Analysis of amino acid sequences of SabHLH169 in S. aralocaspica N-terminal underline indicates bHLH-MYC_N domain; C-terminal underline indicates HLH domain

图3 异子蓬SabHLH169系统进化树及HLH结构域分析 A:SabHLH169系统进化树分析. 系统发育树使用MEGA X软件基于NJ法构建,基于1 000次 bootstrap;Sa:Suaeda aralocaspica,异子蓬;Cq:Chenopodium quinoa,藜麦;Bv:Beta vulgaris,甜菜;At:Arabidopsis thaliana,拟南芥;Nt:Nicotiana tabacum,烟草;So:Spinacia oleracea,菠菜;Os:Oryza sativa,水稻;◆表示异子蓬中的bHLH169。B:SabHLH169与近缘物种的氨基酸序列多重比对。不同颜色背景表示保守程度不同(深蓝>粉红>浅蓝>灰色;黑色背景表示完全保守);上划线表示HLH结构域
Fig. 3 Analysis of the phylogenetic tree and bHLH domain of SabHLH169 in S. aralocaspica A: Analysis of the phylogenetic tree of SabHLH169, which was constructed based on the NJ method using MEGA X software with 1 000 bootstraps. B: Multiple alignment of the bHLH motif of amino acid sequences between SabHLH169 and the closely related species. Different colored letters indicate different degrees of conservation(dark blue > pink red > light blue > grey; the dark background indicates sequence identical); the upper line indicates the HLH domain

图4 异子蓬SabHLH169及其近缘种保守基序分析 A:排名前10的motif碱基富集示意图. 不同颜色方块代表motif1-motif10;字母高度代表该氨基酸出现的频率,高度越高频率越大;B:不同物种bHLH转录因子的保守motif分布. Sa:Suaeda aralocaspica,异子蓬;Cq:Chenopodium quinoa,藜麦;Bv:Beta vulgaris,甜菜;At:Arabidopsis thaliana,拟南芥;Nt:Nicotiana tabacum,烟草,So:Spinacia oleracea,菠菜;Os:Oryza sativa,水稻
Fig. 4 Analysis of the conserved motifs of SabHLH169 in C. quinoa and the closely related species A: Schematic diagram of the base enrichment of the top 10 motifs; boxes in different colors represent motif1-motif10; the height of the letter indicates the frequency of the amino acid, and the higher the height, the greater the frequency. B: Conserved motif analysis of bHLH transcription factors in different species
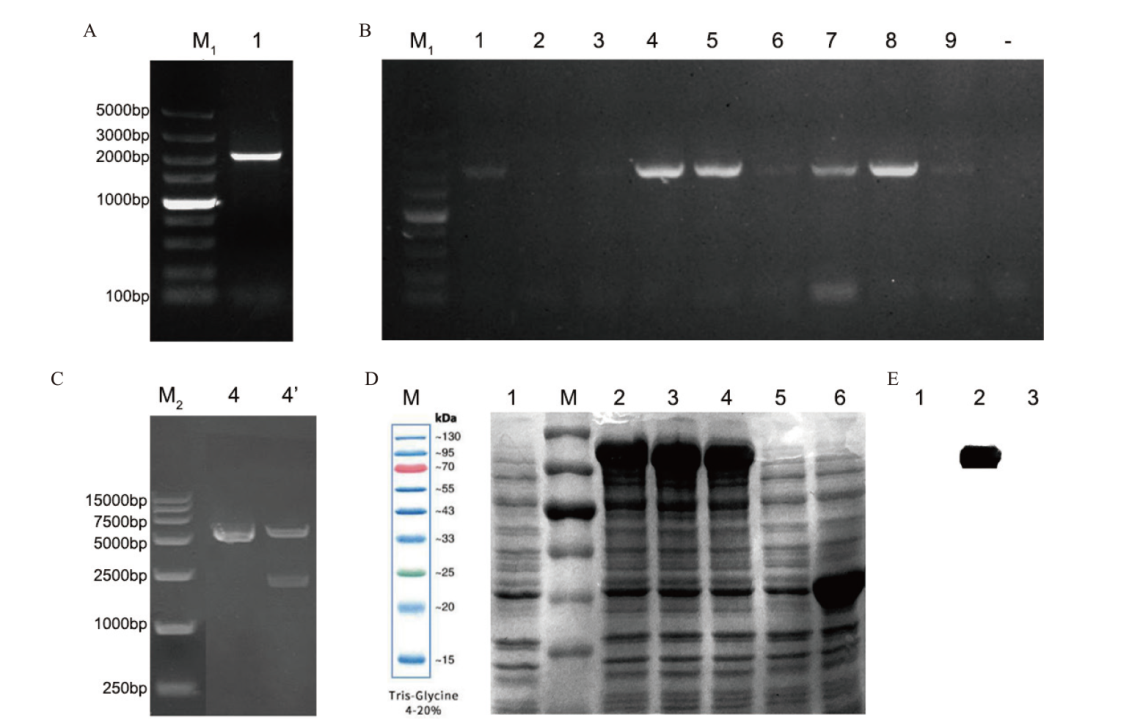
图5 原核表达载体pMal-c2X -SabHLH169的构建及重组蛋白SabHLH169的SDS-PAGE和Western blot分析 A:SabHLH169全长序列的扩增。B:pMal-c2X -SabHLH169的菌液PCR鉴定。-:阴性对照;C:重组质粒的双酶切鉴定。D:SDS-PAGE结果。M:蛋白质Marker;1:pMal-c2X-SabHLH169诱导前;2-4:pMal-c2X-SabHLH169诱导后(0.5、0.8、1.0 mmol/L IPTG);5,6:分别为pMal-c2X诱导前和诱导后;E:Western blot 结果. 1:pMal-c2X -SabHLH169诱导后(0.5 mmol/L IPTG);2:pMal-c2X-SabHLH169诱导前;3:pMal-c2X诱导前
Fig. 5 Construction of procaryotic expression vector pMal-c2X -SabHLH169 and SDS-PAGE and Western blot analysis of SabHLH169 A: Amplification of SabHLH169. B: PCR identification of pMal-c2X-SabHLH169 transformed colonies; -: negative control. C: Double digestion of recombinant plasmid. D: SDS-PAGE results. M: Protein marker; 1: Before induction of pMal-c2X-SabHLH169; 2-4: after induction of pMal-c2X-SabHLH169(0.5 mmol/L, 0.8 mmol/L, 1 mmol/L IPTG); 5, 6: before and after induction of pMal-c2X. E: Western blot results; 1: After induction of pMal-c2X-SabHLH169(0.5 mmol/L IPTG); 2: before induction of pMal-c2X-SabHLH169; 3: before induction of pMal-c2X

图6 植物表达载体pSuper1300-SabHLH169的构建及过表达SabHLH169转基因拟南芥株系的鉴定 A:SabHLH169的扩增;B:pSuper1300-SabHLH169重组菌液PCR鉴定;C:重组质粒的双酶切鉴定;D:过表达SabHLH169转基因拟南芥T0代的鉴定;E,F:3个T1转基因株系OE5、OE16、OE20中SabHLH169的PCR和RT-qPCR 鉴定. +:阳性对照;-:阴性对照;WT:野生型拟南芥;OE5、OE16和OE20:过表达 SabHLH169的3个拟南芥株系;M:DL 5000 DNA marker
Fig. 6 Construction of plant expression vector pSuper1300-SabHLH169 and identification of transgenic A. thaliana with SabHLH169 overexpression A: Amplification of SabHLH169. B: PCR identification of pSuper1300-SabHLH169 transformed colonies. C: Double digestion of recombinant plasmid. D: Identification of the T0 generation of transgenic A. thaliana overexpress SabHLH169. E, F: PCR and RT-qPCR identification of SabHLH169 in three T1 transgenic lines OE5, OE16, OE20. +: positive control; -: negative control. WT: Wild type A. thaliana; OE5, OE16 and OE20: three transgenic A. thaliana lines with SabHLH169 overexpression. M: DL 5000 DNA marker
| 株系 Strain | 阳性苗 Positive seedlings | 阴性苗 Negative seedlings | 卡方检验 Chi-square test(P-value) | 株系 Strain | 阳性苗 Positive seedling | 阴性苗 Negative seedling | 卡方检验 Chi-square test(P-value) | |
|---|---|---|---|---|---|---|---|---|
| OE1 | 134 | 37 | 0.310 | OE13 | 78 | 31 | 0.407 | |
| OE5 | 81 | 26 | 0.867 | OE14 | 64 | 27 | 0.304 | |
| OE6 | 70 | 33 | 0.099 | OE16 | 65 | 26 | 0.431 | |
| OE7 | 79 | 40 | 0.030 | OE17 | 62 | 26 | 0.325 | |
| OE8 | 78 | 32 | 0.322 | OE18 | 54 | 25 | 0.173 | |
| OE11 | 64 | 29 | 0.169 | OE19 | 54 | 26 | 0.121 | |
| OE12 | 61 | 24 | 0.491 | OE20 | 56 | 21 | 0.645 |
表2 SabHLH169 转基因拟南芥T1代进行抗生素筛选3∶1分离比株系
Table 2 Antibiotic screening of SabHLH169 transgenic A. thaliana T1 generation for 3∶1 segregation ratio lines
| 株系 Strain | 阳性苗 Positive seedlings | 阴性苗 Negative seedlings | 卡方检验 Chi-square test(P-value) | 株系 Strain | 阳性苗 Positive seedling | 阴性苗 Negative seedling | 卡方检验 Chi-square test(P-value) | |
|---|---|---|---|---|---|---|---|---|
| OE1 | 134 | 37 | 0.310 | OE13 | 78 | 31 | 0.407 | |
| OE5 | 81 | 26 | 0.867 | OE14 | 64 | 27 | 0.304 | |
| OE6 | 70 | 33 | 0.099 | OE16 | 65 | 26 | 0.431 | |
| OE7 | 79 | 40 | 0.030 | OE17 | 62 | 26 | 0.325 | |
| OE8 | 78 | 32 | 0.322 | OE18 | 54 | 25 | 0.173 | |
| OE11 | 64 | 29 | 0.169 | OE19 | 54 | 26 | 0.121 | |
| OE12 | 61 | 24 | 0.491 | OE20 | 56 | 21 | 0.645 |

图7 甘露醇处理下SabHLH169转基因拟南芥种子萌发实验 A:表型分析;B: 种子萌发率统计
Fig. 7 Seed germination of SabHLH169 transgenic A. thaliana under mannitol treatment A: Phenotype analysis. B: Statistics of seed germination percentage
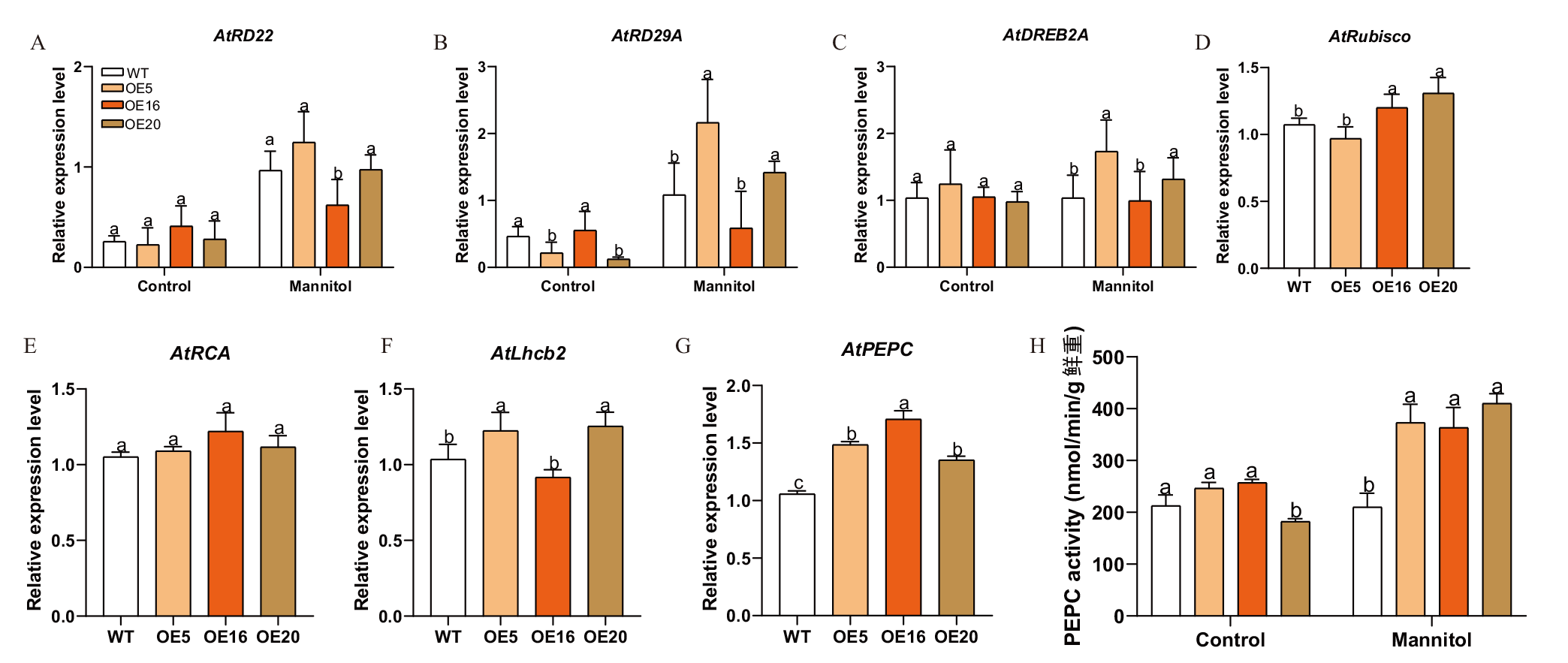
图8 转基因拟南芥干旱和光合作用相关基因的表达及PEPC酶活性分析 A-C:转基因拟南芥中干旱胁迫相关基因的表达分析;D-G:光合作用相关基因的表达分析;H:转基因拟南芥 PEPC 酶活性分析。不同小写字母表示相同条件不同株系之间的基因表达量差异显著(P<0.05)
Fig. 8 Analysis of expression patterns of drought and photosynthesis related genes and PEPC enzyme activity in transgenic A. thaliana A-C: Expressions of drought stress-related genes in transgenic A.thaliana. D-G: Expressions of photosynthesis related genes. H: PEPC enzyme activity in transgenic A. thaliana. Different lowercase letters indicate significant differences in gene expression level at the same condition between different lines(P<0.05)
| [1] |
Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. Plant Cell, 2003, 15(8): 1749-1770.
doi: 10.1105/tpc.013839 pmid: 12897250 |
| [2] |
Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4): 1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [3] |
Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress[J]. Biochemical and Biophysical Research Communications, 2018, 503(2):397-401.
doi: S0006-291X(18)31625-5 pmid: 30057319 |
| [4] |
Castilhos G, Lazzarotto F, Spagnolo-Fonini L, et al. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought[J]. Plant Sci, 2014, 223: 1-7.
doi: 10.1016/j.plantsci.2014.02.010 pmid: 24767109 |
| [5] |
Voznesenskaya EV, Franceschi VR, Kiirats O, et al. Kranz anatomy is not essential for terrestrial C4 plant photosynthesis[J]. Nature, 2001, 414(6863): 543-546.
doi: 10.1038/35107073 |
| [6] |
Edwards GE, Franceschi VR, Voznesenskaya EV. Single-cell C(4)photosynthesis versus the dual-cell(Kranz)paradigm[J]. Annu Rev Plant Biol, 2004, 55: 173-196.
pmid: 15377218 |
| [7] | Cao J, Cheng G, Wang L, et al. Genome-wide identification and analysis of the phosphoenolpyruvate carboxylase gene family in Suaeda aralocaspica, an annual halophyte with single-cellular C4 anatomy[J]. Frontiers in Plant Science, 2021, 12: 1603. |
| [8] | 曹婧. 非花环结构C4盐生植物异子蓬两种PEPC(PEPC-1和PEPC-2)基因的功能及其转录调控研究[D]. 乌鲁木齐: 新疆大学, 2019. |
| Cao J. Function and transcriptional regulation of two types of PEPC(PEPC-1 and PEPC-2)genes in Suaeda aralocaspica, a C4 halophyte with non-Kranz anatomy[D]. Urumqi: Xinjiang University, 2019. | |
| [9] |
Górska AM, Gouveia P, Borba AR, et al. ZmbHLH80 and ZmbHLH90 transcription factors act antagonistically and contribute to regulate PEPC1 cell-specific gene expression in maize[J]. The Plant Journal, 2019, 99(2): 270-285.
doi: 10.1111/tpj.14323 pmid: 30900785 |
| [10] |
Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR[J]. BioTechniques, 2005, 39(4): 519-525.
doi: 10.2144/000112010 pmid: 16235564 |
| [11] |
Zhang TT, Lv W, Zhang HS, et al. Genome-wide analysis of the basic Helix-Loop-Helix(bHLH)transcription factor family in maize[J]. BMC Plant Biol, 2018, 18(1): 235.
doi: 10.1186/s12870-018-1441-z |
| [12] |
Wang PF, Su L, Gao HH, et al. Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis[J]. Front Plant Sci, 2018, 9: 64.
doi: 10.3389/fpls.2018.00064 pmid: 29449854 |
| [13] |
Mao K, Dong Q, Li C, et al. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress[J]. Front Plant Sci, 2017, 8: 480.
doi: 10.3389/fpls.2017.00480 pmid: 28443104 |
| [14] |
Zhao PC, Li XX, Jia JT, et al. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy[J]. J Exp Bot, 2019, 70(1): 269-284.
doi: 10.1093/jxb/ery335 URL |
| [15] |
Wang L, Wang HL, Yin L, et al. Transcriptome assembly in Suaeda aralocaspica to reveal the distinct temporal gene/miRNA alterations between the dimorphic seeds during germination[J]. BMC Genomics, 2017, 18(1): 806.
doi: 10.1186/s12864-017-4209-1 pmid: 29052505 |
| [16] |
Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY[J]. Development, 2007, 134(16): 2959-2968.
pmid: 17626058 |
| [17] |
Wei SW, Xia R, Chen CX, et al. ZmbHLH124 identified in maize recombinant inbred lines contributes to drought tolerance in crops[J]. Plant Biotechnol, 2021, 19(10): 2069-2081.
doi: 10.1111/pbi.v19.10 URL |
| [18] |
Abe H, Yamaguchi-Shinozaki K, Urao T, et al. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression[J]. Plant Cell, 1997, 9(10): 1859-1868.
doi: 10.1105/tpc.9.10.1859 pmid: 9368419 |
| [19] |
Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1[J]. Genetics, 2000, 156(3): 1349-1362.
doi: 10.1093/genetics/156.3.1349 pmid: 11063707 |
| [20] |
Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth[J]. Trends Genet, 1997, 13(4): 152-156.
pmid: 9097726 |
| [21] |
Arnau J, Oliver RP. Inheritance and alteration of transforming DNA during an induced parasexual cycle in the imperfect fungus Cladosporium fulvum[J]. Curr Genet, 1993, 23(5): 508-511.
doi: 10.1007/BF00312643 URL |
| [22] |
Provvidenti R, Gonsalves D. Inheritance of resistance to cucumber mosaic virus in a transgenic tomato line expressing the coat protein gene of the white leaf strain[J]. J Hered, 1995, 86(2): 85-88.
doi: 10.1093/oxfordjournals.jhered.a111553 URL |
| [23] | 贾庆利. 外源基因在拟南芥转基因植株中的遗传研究[D]. 杨凌: 西北农林科技大学, 2004. |
| Jia QL. Genetic study of exogenous genes in transgenic Arabidopsis plants[D]. Yangling: Northwest A & F University, 2004. | |
| [24] |
Helm LT, Shi HY, Lerdau MT, et al. Solar-induced chlorophyll fluorescence and short-term photosynthetic response to drought[J]. Ecol Appl, 2020, 30(5): e02101.
doi: 10.1002/eap.v30.5 URL |
| [25] |
Sajad Majeed Z, Nancy G, Muslima N, et al. Impact of drought on photosynthesis: Molecular perspective[J]. Plant Gene, 2017, 11(2): 154-159.
doi: 10.1016/j.plgene.2017.04.003 URL |
| [26] |
Spreitzer RJ, Salvucci ME. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme[J]. Annu Rev Plant Biol, 2002, 53: 449-475.
pmid: 12221984 |
| [27] | Tissue DT, Thomas RB, Strain BR. Long-term effects of elevated CO2 and nutrients on photosynthesis and rubisco in loblolly pine seedlings[J]. Plant Cell & Environment, 2010, 16(7): 859-865. |
| [28] |
Kurek I, Chang TK, Bertain SM, et al. Enhanced Thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress[J]. Plant Cell, 2007, 19(10): 3230-3241.
doi: 10.1105/tpc.107.054171 URL |
| [29] |
Demirevska-Kepova K, Holzer R, Simova-Stoilova L, et al. Heat stress effects on ribulose-1, 5-bisphosphate carboxylase/oxygenase, Rubisco binding protein and Rubisco activase in wheat leaves[J]. Biol Plant, 2005, 49(4): 521-525.
doi: 10.1007/s10535-005-0045-2 URL |
| [30] |
Yin ZT, Zhang ZL, Deng DX, et al. Characterization of Rubisco activase genes in maize: an α-isoform gene functions alongside a β-isoform gene[J]. Plant Physiol, 2014, 164(4): 2096-2106.
doi: 10.1104/pp.113.230854 pmid: 24510763 |
| [31] |
Sharkey TD, Badger MR, von Caemmerer S, et al. Increased heat sensitivity of photosynthesis in tobacco plants with reduced Rubisco activase[J]. Photosynth Res, 2001, 67(1): 147-156.
doi: 10.1023/A:1010633823747 URL |
| [32] |
Wang D, Li XF, Zhou ZJ, et al. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant[J]. Physiol Plant, 2010, 139(1): 55-67.
doi: 10.1111/j.1399-3054.2009.01344.x pmid: 20059735 |
| [33] | Jackowski G, Kacprzak K, Jansson S. Identification of Lhcb1/Lhcb2/Lhcb3 heterotrimers of the main light-harvesting chlorophyll a/b-protein complex of Photosystem II(LHC II)[J]. Biochim Biophys Acta BBA Bioenerg, 2001, 1504(2/3): 340-345. |
| [34] |
O’Leary B, Park J, Plaxton WC. The remarkable diversity of plant PEPC(phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs[J]. Biochem J, 2011, 436(1): 15-34.
doi: 10.1042/BJ20110078 URL |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [3] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [4] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [7] | 丁凯鑫, 王立春, 田国奎, 王海艳, 李凤云, 潘阳, 庞泽, 单莹. 烯效唑缓解植物干旱损伤的研究进展[J]. 生物技术通报, 2023, 39(6): 1-11. |
| [8] | 王春语, 李政君, 王平, 张丽霞. 高粱表皮蜡质缺失突变体sb1抗旱生理生化分析[J]. 生物技术通报, 2023, 39(5): 160-167. |
| [9] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [10] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [11] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [12] | 于波, 秦晓惠, 赵杨. 植物感应干旱信号的机制[J]. 生物技术通报, 2023, 39(11): 6-17. |
| [13] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [14] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [15] | 安昌, 陆琳, 沈梦千, 陈盛圳, 叶康卓, 秦源, 郑平. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||