








生物技术通报 ›› 2022, Vol. 38 ›› Issue (10): 216-225.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0099
张泽颖1,3( ), 范清锋1,3, 邓云峰1,3, 韦廷舟1,3, 周正富2, 周建1,3, 王劲2, 江世杰1,3(
), 范清锋1,3, 邓云峰1,3, 韦廷舟1,3, 周正富2, 周建1,3, 王劲2, 江世杰1,3( )
)
收稿日期:2021-01-21
出版日期:2022-10-26
发布日期:2022-11-11
作者简介:张泽颖,女,研究方向:微生物酶工程;E-mail:基金资助:
ZHANG Ze-ying1,3( ), FAN Qing-feng1,3, DENG Yun-feng1,3, WEI Ting-zhou1,3, ZHOU Zheng-fu2, ZHOU Jian1,3, WANG Jin2, JIANG Shi-jie1,3(
), FAN Qing-feng1,3, DENG Yun-feng1,3, WEI Ting-zhou1,3, ZHOU Zheng-fu2, ZHOU Jian1,3, WANG Jin2, JIANG Shi-jie1,3( )
)
Received:2021-01-21
Published:2022-10-26
Online:2022-11-11
摘要:
琼氏不动杆菌WCO-9是从油污污染的土壤中分离的一株高产脂肪酶菌株,所产脂肪酶具有显著优于对照同种菌株ATCC 17908的水解活性。为探究WCO-9菌株高效产酶机制,挖掘新型特异脂肪酶功能基因,采用Oxford Nanopore技术完成WCO-9菌株的全基因组测序,并对同属菌株进行比较基因组学分析。结果显示,脂肪酶产生菌WCO-9的基因组大小为3.19 Mb,GC含量38.62%,含有2 929个编码基因,其中包含11个脂肪酶编码基因(1个特异基因)和3个脂肪酶分子伴侣基因;共线性分析也说明WCO-9菌株基因组与对照菌株ATCC 17908具有显著差异。WCO-9菌株全基因组序列分析,对该菌的高效产酶机制研究及新型脂肪酶特异基因挖掘具有重要意义,为后期高产脂肪酶工程菌的创制及工业化应用提供理论基础。
张泽颖, 范清锋, 邓云峰, 韦廷舟, 周正富, 周建, 王劲, 江世杰. 一株高产脂肪酶菌株WCO-9全基因组测序及比较基因组分析[J]. 生物技术通报, 2022, 38(10): 216-225.
ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9[J]. Biotechnology Bulletin, 2022, 38(10): 216-225.
| 菌株Strain | 脂肪酶活力Lipase activity/(U·L-1) |
|---|---|
| WCO-9 | 2 833.33±166.67A |
| ATCC 17908 | 24.44±8.39B |
表1 菌株WCO-9所产脂肪酶对底物橄榄油的水解能力分析
Table 1 Analysis of the hydrolysis ability of lipase from WCO-9 strain on the substrate olive oil by GB/T 23535-2009 method
| 菌株Strain | 脂肪酶活力Lipase activity/(U·L-1) |
|---|---|
| WCO-9 | 2 833.33±166.67A |
| ATCC 17908 | 24.44±8.39B |
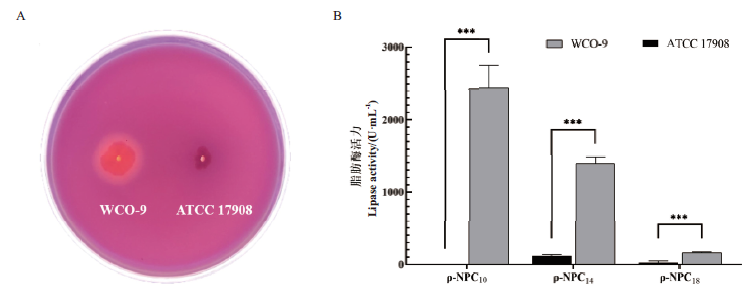
图 2 WCO-9菌株产脂肪酶活性分析 A:罗丹明B-油脂同化平板法分析WCO-9菌株产脂肪酶能力;B:WCO-9菌株脂肪酶水解不同对硝基苯酯的活性差异,***P<0.01
Fig. 2 Lipase activity analysis of WCO-9 strain A: Analysis of lipase-producing ability of WCO-9 strain by rhodamine B-oil assimilation plate method; B: Differences in hydrolytic activity of lipase from WCO-9 strain towards various ρ-NP esters, ***P<0.01
| 编号No. | 通路Pathway ID | 功能描述Description | 基因编号Gene ID |
|---|---|---|---|
| 1 | ko00071 | 脂肪酸降解 Fatty acid degradation | GE000138、GE000223、GE000309、GE000310、GE000604、GE000605、GE000616、GE001181、GE001341、GE001407、GE001474、GE001518、GE001530、GE001747、GE001825、GE001884、GE001964 |
| 2 | ko00561 | 甘油酯代谢 Glycerolipid metabolism | GE000861、GE000871、GE001701、GE001702、GE002495、GE002594 |
| 3 | ko00564 | 甘油磷脂代谢 Glycerophospholipid metabolism | GE000081、GE000318、GE000487、GE000575、GE000725、GE000800、GE000863、GE000871、GE001133、GE001134、GE001196、GE001398、GE001531、GE001566、GE002494、GE002495、GE002594、GE002873、GE002885 |
| 4 | ko01212 | 脂肪酸代谢 Fatty acid metabolism | GE000138、GE000223、GE000309、GE000310、GE000573、GE000587、GE000604、GE000605、GE000616、GE001181、GE001182、GE001359、GE001360、GE001404、GE001474、GE001769、GE001820、GE001825、GE002165、GE002197、GE002198、GE002362、GE002463、GE002920 |
表2 产脂肪酶菌株WCO-9基因组脂肪代谢降解通路及相关基因
Table 2 Fat metabolism and degradation pathways in lipase-producing strain WCO-9 genome and their related genes
| 编号No. | 通路Pathway ID | 功能描述Description | 基因编号Gene ID |
|---|---|---|---|
| 1 | ko00071 | 脂肪酸降解 Fatty acid degradation | GE000138、GE000223、GE000309、GE000310、GE000604、GE000605、GE000616、GE001181、GE001341、GE001407、GE001474、GE001518、GE001530、GE001747、GE001825、GE001884、GE001964 |
| 2 | ko00561 | 甘油酯代谢 Glycerolipid metabolism | GE000861、GE000871、GE001701、GE001702、GE002495、GE002594 |
| 3 | ko00564 | 甘油磷脂代谢 Glycerophospholipid metabolism | GE000081、GE000318、GE000487、GE000575、GE000725、GE000800、GE000863、GE000871、GE001133、GE001134、GE001196、GE001398、GE001531、GE001566、GE002494、GE002495、GE002594、GE002873、GE002885 |
| 4 | ko01212 | 脂肪酸代谢 Fatty acid metabolism | GE000138、GE000223、GE000309、GE000310、GE000573、GE000587、GE000604、GE000605、GE000616、GE001181、GE001182、GE001359、GE001360、GE001404、GE001474、GE001769、GE001820、GE001825、GE002165、GE002197、GE002198、GE002362、GE002463、GE002920 |
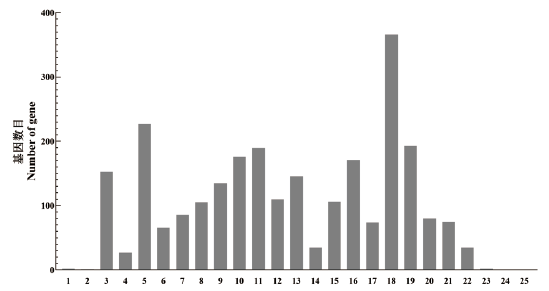
图6 产脂肪酶菌株WCO-9 COG功能分类 1:RNA加工和修饰;2:染色质结构和动力学;3:能源生产和转换;4:细胞周期控制、细胞分裂、染色体分割;5:氨基酸转运与代谢;6:核苷酸转运和代谢;7:碳水化合物运输和代谢;8:辅酶转运和代谢;9:脂质转运与代谢;10:翻译、核糖体结构和生物起源;11:转录;12:复制、重组和修复;13:细胞壁/膜/被膜生物发生;14:细胞运动;15:翻译后修饰、蛋白质周转、分子伴侣;16:无机离子转运与代谢;17:次级代谢物生物合成、运输和分解代谢;18:一般功能预测;19:功能未知;20:信号转导机制;21:细胞内运输、分泌和囊泡运输;22:防御机制;23:胞外结构;24:核结构;25:细胞骨架
Fig. 6 COG function classification chart of lipase-produc-ing strain WCO-9 1:RNA processing and modification. 2:Chromatin structure and dynamics. 3:Energy production and conversion. 4:Cell cycle control,cell division,chromosome partitioning. 5:Amino acid transport and metabolism. 6:Nucleotide transport and metabolism. 7:Carbohydrate transport and metabolism. 8:Coenzyme transport and metabolism. 9:Lipid transport and metabolism. 10:Translation,ribosomal structure and biogenesis. 11:Transcription. 12:Replication,recombination and repair. 13:Cell wall/membrane/envelope biogenesis. 14:Cell motility. 15:Posttranslational modification,protein turnover,chaperones. 16:Inorganic ion transport and metabolism. 17:Secondary metabolites biosynthesis,transport and catabolism. 18:General function prediction only. 19:Function unknown. 20:Signal transduction mechanisms. 21:Intracellular trafficking,secretion,and vesicular transport. 22:Defense mechanisms. 23:Extracellular structures. 24:Nuclear structure. 25:Cytoskeleton
| 类型Type | 基因组大小Genome size /bp | GC含量GC content /% | 编码基因数目Coding genes | GenBank登录号GenBank INSDC ID |
|---|---|---|---|---|
| WCO-9 | 3 193 903 | 38.47 | 2 943 | CP090890.1 |
| Acinetobacter junii ATCC 17908 | 3 357 744 | 38.90 | 3 185 | APPX00000000.1 |
| Acinetobacter baumannii ATCC 19606 | 3 927 723 | 39.18 | 3 681 | CP058289.1 |
| Acinetobacter chinensis WCHAc010005 | 3 600 635 | 42.48 | 3 434 | CP032134.1 |
表3 菌株WCO-9与3株不动杆菌全基因组序列基本特征比较
Table 3 Comparison of basic characteristics of whole genome sequences of strain WCO-9 and 3 strains of Acinetobacter
| 类型Type | 基因组大小Genome size /bp | GC含量GC content /% | 编码基因数目Coding genes | GenBank登录号GenBank INSDC ID |
|---|---|---|---|---|
| WCO-9 | 3 193 903 | 38.47 | 2 943 | CP090890.1 |
| Acinetobacter junii ATCC 17908 | 3 357 744 | 38.90 | 3 185 | APPX00000000.1 |
| Acinetobacter baumannii ATCC 19606 | 3 927 723 | 39.18 | 3 681 | CP058289.1 |
| Acinetobacter chinensis WCHAc010005 | 3 600 635 | 42.48 | 3 434 | CP032134.1 |
| WCO-9基因编号 Gene ID of WCO-9 | 功能注释 Function annotation | 注释来源数据库 Source of function | GenBank 中序列一致性 Sequence identity in GenBank/% | 同源菌株 Homologous source strain | ATCC 17908中同源蛋白编号 Homologous protein ID of ATCC 17908 | ATCC 17908中同源蛋白序列一致性Sequence identity with homologous protein ID of ATCC 17908/% |
|---|---|---|---|---|---|---|
| GE000005 | 脂肪酶2 Lipase 2 | Swissprot、TrEMBL | 100 | Acinetobacter junii NIPH 182 | WP_004965292.1 | 99.44 |
| GE000880 | 脂肪酶1 Lipase 1 | Swissprot、TrEMBL | 100 | Acinetobacter junii NBRC 110497 | WP_004963343.1 | 99.41 |
| GE000882 | GDSL脂肪酶 Lipase_GDSL | Pfam | 100 | Acinetobacter junii KCTC 42611 | ENV66375.1 | 99.52 |
| GE001015 | 甘油三酯脂肪酶活性 Triglyceride lipase activity | GO | 99.78 | Acinetobacter junii YR7 | ENV66472.1 | 84.53 |
| GE001547 | GDSL脂肪酶2 Lipase GDSL 2 | Pfam | 99.16 | Acinetobacter junii KCTC 42611 | WP_004962046.1 | 96.22 |
| GE001701 | 三酰甘油脂肪酶 Triacylglycerol lipase | KEGG | 100 | Acinetobacter junii NIPH 182 | ENV65669.1 | 42.90 |
| GE001702 | 三酰甘油脂肪酶 Triacylglycerol lipase | KEGG | 100 | Acinetobacter junii NIPH 182 | WP_035333945.1 | 40.29 |
| GE001718 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 100 | Acinetobacter junii TUM15553 | WP_004962461.1 | 98.78 |
| GE001892 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 100 | Acinetobacter junii SH205 | WP_004953257.1 | 99.80 |
| GE001931 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 90.76 | Acinetobacter sp. SAT183 | — | — |
| GE002660 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 99.38 | Acinetobacter junii NIPH 182 | ENV65669.1 | 78.72 |
| GE000502 | 脂肪酶伴侣 Lipase chaperone | Pfam | 99.67 | Acinetobacter junii SB132 | — | — |
| GE002659 | 脂肪酶伴侣 Lipase chaperone | Pfam、Swissprot、 TrEMBL | 98.56 | Acinetobacter junii YR7 | WP_004964537.1 | 63.98 |
| GE001700 | 脂肪酶伴侣 Lipase chaperone | Pfam、Swissprot、 TrEMBL | 100 | Acinetobacter junii lzh-X15 | WP_004964537.1 | 32.56 |
表4 脂肪酶基因与NCBI数据库中基因序列相似度
Table 4 Sequence similarity between lipase genes and genes in NCBI database
| WCO-9基因编号 Gene ID of WCO-9 | 功能注释 Function annotation | 注释来源数据库 Source of function | GenBank 中序列一致性 Sequence identity in GenBank/% | 同源菌株 Homologous source strain | ATCC 17908中同源蛋白编号 Homologous protein ID of ATCC 17908 | ATCC 17908中同源蛋白序列一致性Sequence identity with homologous protein ID of ATCC 17908/% |
|---|---|---|---|---|---|---|
| GE000005 | 脂肪酶2 Lipase 2 | Swissprot、TrEMBL | 100 | Acinetobacter junii NIPH 182 | WP_004965292.1 | 99.44 |
| GE000880 | 脂肪酶1 Lipase 1 | Swissprot、TrEMBL | 100 | Acinetobacter junii NBRC 110497 | WP_004963343.1 | 99.41 |
| GE000882 | GDSL脂肪酶 Lipase_GDSL | Pfam | 100 | Acinetobacter junii KCTC 42611 | ENV66375.1 | 99.52 |
| GE001015 | 甘油三酯脂肪酶活性 Triglyceride lipase activity | GO | 99.78 | Acinetobacter junii YR7 | ENV66472.1 | 84.53 |
| GE001547 | GDSL脂肪酶2 Lipase GDSL 2 | Pfam | 99.16 | Acinetobacter junii KCTC 42611 | WP_004962046.1 | 96.22 |
| GE001701 | 三酰甘油脂肪酶 Triacylglycerol lipase | KEGG | 100 | Acinetobacter junii NIPH 182 | ENV65669.1 | 42.90 |
| GE001702 | 三酰甘油脂肪酶 Triacylglycerol lipase | KEGG | 100 | Acinetobacter junii NIPH 182 | WP_035333945.1 | 40.29 |
| GE001718 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 100 | Acinetobacter junii TUM15553 | WP_004962461.1 | 98.78 |
| GE001892 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 100 | Acinetobacter junii SH205 | WP_004953257.1 | 99.80 |
| GE001931 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 90.76 | Acinetobacter sp. SAT183 | — | — |
| GE002660 | 甘油三酯脂肪酶 Triglyceride lipase activity | GO | 99.38 | Acinetobacter junii NIPH 182 | ENV65669.1 | 78.72 |
| GE000502 | 脂肪酶伴侣 Lipase chaperone | Pfam | 99.67 | Acinetobacter junii SB132 | — | — |
| GE002659 | 脂肪酶伴侣 Lipase chaperone | Pfam、Swissprot、 TrEMBL | 98.56 | Acinetobacter junii YR7 | WP_004964537.1 | 63.98 |
| GE001700 | 脂肪酶伴侣 Lipase chaperone | Pfam、Swissprot、 TrEMBL | 100 | Acinetobacter junii lzh-X15 | WP_004964537.1 | 32.56 |
| [1] |
Javed S, Azeem F, Hussain S, et al. Bacterial lipases:a review on purification and characterization[J]. Prog Biophys Mol Biol, 2018, 132:23-34.
doi: 10.1016/j.pbiomolbio.2017.07.014 URL |
| [2] |
Jung J, Park W. Acinetobacter species as model microorganisms in environmental microbiology:current state and perspectives[J]. Appl Microbiol Biotechnol, 2015, 99(6):2533-2548.
doi: 10.1007/s00253-015-6439-y URL |
| [3] | Ken Ugo A, Vivian Amara A, Cn I, et al. Microbial lipases:a prospect for biotechnological industrial catalysis for green products:a review[J]. Ferment Technol, 2017, 6(2):1000144. |
| [4] | Chandra P, Enespa, Singh DP. Microplastic degradation by bacteria in aquatic ecosystem[M]//Chowdhary P, Raj A, Verma D, et al. Microorganisms for Sustainable Environment and Health. Amsterdam:Elsevier, 2020:431-467. |
| [5] |
Filho DG, Silva AG, Guidini CZ. Lipases:sources, immobilization methods, and industrial applications[J]. Appl Microbiol Biotechnol, 2019, 103(18):7399-7423.
doi: 10.1007/s00253-019-10027-6 URL |
| [6] |
Sarmah N, Revathi D, Sheelu G, et al. Recent advances on sources and industrial applications of lipases[J]. Biotechnol Prog, 2018, 34(1):5-28.
doi: 10.1002/btpr.2581 URL |
| [7] |
Vanleeuw E, Winderickx S, Thevissen K, et al. Substrate-specificity of Candida rugosa lipase and its industrial application[J]. ACS Sustain Chem Eng, 2019, 7(19):15828-15844.
doi: 10.1021/acssuschemeng.9b03257 |
| [8] |
黄阳天, 陆育彪, 黄益帖, 等. 产电海洋脂肪酶生产菌的分离筛选鉴定及培养条件研究[J]. 生物技术通报, 2020, 36(12):91-97.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0833 |
| Huang YT, Lu YB, Huang YT, et al. Screening and identification of marine electricity-producing and lipase-producing bacteria and preliminary study on its culture conditions[J]. Biotechnol Bull, 2020, 36(12):91-97. | |
| [9] |
Haq A, Adeel S, Khan A, et al. Screening of lipase-producing bacteria and optimization of lipase-mediated biodiesel production from Jatropha curcas seed oil using whole cell approach[J]. Bioenergy Res, 2020, 13(4):1280-1296.
doi: 10.1007/s12155-020-10156-1 URL |
| [10] |
Ng PC, Kirkness EF. Whole genome sequencing[J]. Methods Mol Biol, 2010, 628:215-226.
doi: 10.1007/978-1-60327-367-1_12 pmid: 20238084 |
| [11] | 吕瑞瑞, 李伟程, 康小红, 等. 副干酪乳杆菌PC-01全基因组测序及不同副干酪乳杆菌菌株比较基因组学分析[J]. 微生物学通报, 2021(9):3025-3038. |
| Lv RR, Li WC, Kang XH, et al. Whole genome sequencing of Lactobacillus paracasei PC-01 and comparative genomics analysis about Lactobacillus paracasei strains[J]. J Chin Inst Food Sci Technol, 2021(9):3025-3038. | |
| [12] | Lim S, Chang DH, Kim BC. Whole-genome sequence of Bacillus solimangrovi GH 2-4 T, isolated from mangrove soil[J]. Genom Data, 2016, 10:89-90. |
| [13] |
Patel RK, Shah RK, Prajapati VS, et al. Draft genome analysis of Acinetobacter indicus strain UBT1, an efficient lipase and biosurfactant producer[J]. Curr Microbiol, 2021, 78(4):1238-1244.
doi: 10.1007/s00284-021-02380-5 URL |
| [14] |
Bouvet PJM, Grimont PAD. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii[J]. Int J Syst Bacteriol, 1986, 36(2):228-240.
doi: 10.1099/00207713-36-2-228 URL |
| [15] | 徐伟芳, 黄涛杨, 周敏, 等. 一株脂肪酶产生菌的筛选鉴定及其酶学性质研究[J]. 西南大学学报:自然科学版, 2017, 39(5):62-69. |
| Xu WF, Huang TY, Zhou M, et al. Research on screening and identification of a lipase-producing bacterial strain and its enzymatic properties[J]. J Southwest Univ Nat Sci Ed, 2017, 39(5):62-69. | |
| [16] |
Han RH, Lee JE, Yoon SH, et al. Acinetobacter pullicarnis sp. nov. isolated from chicken meat[J]. Arch Microbiol, 2020, 202(4):727-732.
doi: 10.1007/s00203-019-01785-y URL |
| [17] |
Elnar AG, Kim MG, Lee JE, et al. Acinetobacter pullorum sp. nov., isolated from chicken meat[J]. J Microbiol Biotechnol, 2020, 30(4):526-532.
doi: 10.4014/jmb.2002.02033 URL |
| [18] |
Carvalheira A, Gonzales-Siles L, Salvà-Serra F, et al. Acinetobacter portensis sp. nov. and Acinetobacter guerrae sp. nov., isolated from raw meat[J]. Int J Syst Evol Microbiol, 2020, 70(8):4544-4554.
doi: 10.1099/ijsem.0.004311 URL |
| [19] |
Qin JY, Feng Y, Lü XJ, et al. Characterization of Acinetobacter chengduensis sp. nov., isolated from hospital sewage and capable of acquisition of carbapenem resistance genes[J]. Syst Appl Microbiol, 2020, 43(4):126092.
doi: 10.1016/j.syapm.2020.126092 URL |
| [20] | Zhu WT, Dong K, Yang J, et al. Acinetobacter lanii sp. nov., Acinetobacter shaoyimingii sp. nov. and Acinetobacter wanghuae sp. nov., isolated from faeces of Equus kiang[J]. Int J Syst Evol Microbiol, 2021, 71(1):004567. |
| [21] |
Acer Ö, Güven K, Poli A, et al. Acinetobacter mesopotamicus sp. nov., petroleum-degrading bacterium, isolated from petroleum-contaminated soil in Diyarbakir, in the southeast of Turkey[J]. Curr Microbiol, 2020, 77(10):3192-3200.
doi: 10.1007/s00284-020-02134-9 URL |
| [22] |
Rivera-Pérez C, de los Ángeles Navarrete del Toro M, García-Carreño F. Purification and characterization of an intracellular lipase from pleopods of whiteleg shrimp(Litopenaeus vannamei)[J]. Comp Biochem Physiol B Biochem Mol Biol, 2011, 158(1):99-105.
doi: 10.1016/j.cbpb.2010.10.004 URL |
| [23] |
Grbavčić S, Bezbradica D, Izrael-Živković L, et al. Production of lipase and protease from an indigenous Pseudomonas aeruginosa strain and their evaluation as detergent additives:compatibility study with detergent ingredients and washing performance[J]. Bioresour Technol, 2011, 102(24):11226-11233.
doi: 10.1016/j.biortech.2011.09.076 URL |
| [24] |
Fetzner S, Steiner RA. Cofactor-independent oxidases and oxygenases[J]. Appl Microbiol Biotechnol, 2010, 86(3):791-804.
doi: 10.1007/s00253-010-2455-0 pmid: 20157809 |
| [25] |
Kapoor M, Gupta MN. Lipase promiscuity and its biochemical applications[J]. Process Biochem, 2012, 47(4):555-569.
doi: 10.1016/j.procbio.2012.01.011 URL |
| [26] |
Snellman EA, Sullivan ER, Colwell RR. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1[J]. Eur J Biochem, 2002, 269(23):5771-5779.
pmid: 12444965 |
| [27] |
Zheng XM, Wu NF, Fan YL. Characterization of a novel lipase and its specific foldase from Acinetobacter sp. XMZ-26[J]. Process Biochem, 2012, 47(4):643-650.
doi: 10.1016/j.procbio.2012.01.005 URL |
| [28] | 桑鹏, 刘林波, 陈贵元, 等. 大理弥渡热泉耐热脂肪酶产生菌的筛选及其酶活性研究[J]. 中国饲料, 2020(3):27-31. |
| Sang P, Liu LB, Chen GY, et al. Isolation and identification of a strain producing thermostable lipase and studying on its enzymatic properties[J]. China Feed, 2020(3):27-31. | |
| [29] |
陈体强, 徐晓兰, 石林春, 等. 紫芝栽培品种‘武芝2号’(‘紫芝S2’)全基因组测序及分析[J]. 生物技术通报, 2021, 37(11):42-56.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1158 |
| Chen TQ, Xu XL, Shi LC, et al. Sequencing and analysis of the whole genome of Zizhi cultivar ‘Wuzhi No. 2’(Ganoderma sp. strain Zizhi S2)[J]. Biotechnol Bull, 2021, 37(11):42-56. | |
| [30] | Singh M, de Silva PM, Al-Saadi Y, et al. Characterization of extremely drug-resistant and hypervirulent Acinetobacter baumannii AB030[J]. Antibiotics(Basel), 2020, 9(6):328. |
| [31] |
Darling ACE, Mau B, Blattner FR, et al. Mauve:multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Res, 2004, 14(7):1394-1403.
pmid: 15231754 |
| [1] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [2] | 陈金行, 张逸, 张军涛, 未本美, 王宏勋, 郑明明. 固定化脂肪酶的创制及其在乙酸肉桂酯无溶剂制备中的应用[J]. 生物技术通报, 2023, 39(9): 97-104. |
| [3] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [4] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [5] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| [6] | 和梦颖, 刘文彬, 林震鸣, 黎尔彤, 汪洁, 金小宝. 一株抗革兰阳性菌的戈登氏菌WA4-43全基因组测序与分析[J]. 生物技术通报, 2023, 39(2): 232-242. |
| [7] | 张傲洁, 李青云, 宋文红, 颜少慧, 唐爱星, 刘幽燕. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303. |
| [8] | 王帅, 吕鸿睿, 张昊, 吴占文, 肖翠红, 孙冬梅. 解磷菌PSB-R全基因组测序鉴定及其解磷特性分析[J]. 生物技术通报, 2023, 39(1): 274-283. |
| [9] | 薛清, 杜虹锐, 薛会英, 王译浩, 王暄, 李红梅. 苜蓿滑刃线虫线粒体基因组及其系统发育研究[J]. 生物技术通报, 2021, 37(7): 98-106. |
| [10] | 吴蓉, 曹佳睿, 曹君, 刘飞翔, 杨猛, 苏二正. 南极假丝酵母脂肪酶B基因在大肠杆菌中的表达和发酵优化[J]. 生物技术通报, 2021, 37(2): 138-148. |
| [11] | 陈体强, 徐晓兰, 石林春, 钟礼义. 紫芝栽培品种‘武芝2号’(‘紫芝S2’)全基因组测序及分析[J]. 生物技术通报, 2021, 37(11): 42-56. |
| [12] | 郭鹤宝, 王星, 何山文, 张晓霞. 表型特征结合基因组分析鉴定不同菌落形态Bacillus velezensis ACCC 19742[J]. 生物技术通报, 2020, 36(2): 142-148. |
| [13] | 黄阳天, 陆育彪, 黄益帖, 孟繁龙, 徐凯文, 李鹏. 产电海洋脂肪酶生产菌的分离筛选鉴定及培养条件研究[J]. 生物技术通报, 2020, 36(12): 91-97. |
| [14] | 朱彩林, 吕祥, 夏小乐. 盖子区域氨基酸的定点突变对T1脂肪酶酶学性质的影响[J]. 生物技术通报, 2020, 36(11): 94-102. |
| [15] | 蔡玉贞, 白巧燕, 苏敏, 唐良华. 脂肪酶底物结合口袋的分子改造策略及进展[J]. 生物技术通报, 2020, 36(11): 173-180. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||