








生物技术通报 ›› 2022, Vol. 38 ›› Issue (12): 300-311.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0098
盛雪晴( ), 赵楠, 林亚秋, 陈定双, 王瑞龙, 李傲, 王永, 李艳艳(
), 赵楠, 林亚秋, 陈定双, 王瑞龙, 李傲, 王永, 李艳艳( )
)
收稿日期:2022-01-21
出版日期:2022-12-26
发布日期:2022-12-29
作者简介:盛雪晴,女,研究方向:动物遗传育种;E-mail:基金资助:
SHENG Xue-qing( ), ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan(
), ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan( )
)
Received:2022-01-21
Published:2022-12-26
Online:2022-12-29
摘要:
以成年健康简州大耳羊为实验动物,利用RT-PCR技术克隆山羊ZNF32基因。利用生物信息学方法对其进行表达特性分析,并利用实时荧光定量PCR(real-time quantitative PCR,qPCR)技术检测ZNF32在山羊各组织中和诱导分化不同阶段的皮下脂肪细胞中的表达水平。同时构建过表达载体并利用qPCR技术检测过表达或干扰ZNF32对增殖抑制相关基因表达的影响,探究其对山羊皮下前体脂肪细胞增殖的作用。结果显示,克隆所得山羊ZNF32核苷酸序列长度为1 049 bp,其中CDS区长为822 bp,编码273个氨基酸;ZNF32分子量为30 983.03 Da,理论等电点为9.52,亲水性平均值为-0.813;在氨基酸序列组成中,丝氨酸含量为9.2%,占比最高;蛋白质二级结构预测显示,149个氨基酸形成无规卷曲,含量最高;氨基酸同源性比对结果显示,简州大耳羊与绵羊氨基酸序列相似性最高且在进化上亲缘关系最近;组织表达谱表明,ZNF32基因在山羊各个组织中广泛表达,其中在大肠中表达量最高(P<0.01);时序表达谱表明,ZNF32基因表达水平呈上升趋势,在96 h表达量达到最高,极显著高于分化前(P<0.01);qPCR技术检测结果表明,过表达ZNF32基因抑制增殖抑制相关基因p27和p57的表达,干扰ZNF32基因促进p21、p27、p53及p57的表达。以上结果表明,ZNF32基因可能是山羊皮下脂肪细胞分化和增殖的正调控因子。
盛雪晴, 赵楠, 林亚秋, 陈定双, 王瑞龙, 李傲, 王永, 李艳艳. 山羊ZNF32的克隆及表达分析[J]. 生物技术通报, 2022, 38(12): 300-311.
SHENG Xue-qing, ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan. Cloning and Expression Analysis of ZNF32 Gene in Goat[J]. Biotechnology Bulletin, 2022, 38(12): 300-311.
| 目的基因 Target gene | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 产物长度 Product length/bp | 用途 Purpose |
|---|---|---|---|---|
| ZNF32 | S:CCGCAAGGGTCTAGCTG | 58 | 1 049 | 克隆 |
| A:TGGATAGGCTGTTTAATCTCAGATG | Clone | |||
| ZNF32 | S:TGGAAGATATGCCCAGAATGTAG | 60 | 293 | qPCR |
| A:CACAATGGGTACACTCGGG | ||||
| ZNF32 | S:CCGGAATTCCATGTTTGGATTTCCAACAGCTACCC | 58 | 822 | 过表达载体构建 |
| A:CCGCTCGAGACTTACAGGGTGAGCCTTTGTGAGC | Overexpression plasmid construction | |||
| Si-NC | S:UUCUCCGAACGUGUCACGUTT | 60 | 干扰 | |
| A:ACGUGACACGUUCGGAGAATT | Interference | |||
| Si-ZNF32 | S:GGGUAGUCUAACAUUACAUTT | 60 | 干扰 | |
| A:AUGUAAUGUUAGACUACCCTT | Interference | |||
| TBP | S:AACAGCCTCCCACCTTATGC | 60 | 155 | 内参 |
| A:TGCTGCTCCTCCTAGAC | Reference | |||
| UXT | S:GCAAGTGGATTTGGGCTGTAAC | 60 | 180 | 内参 |
| A:ATGGAGTCCTTGGTGAGGTTGT | Reference |
表1 引物信息
Table 1 Primers information
| 目的基因 Target gene | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 产物长度 Product length/bp | 用途 Purpose |
|---|---|---|---|---|
| ZNF32 | S:CCGCAAGGGTCTAGCTG | 58 | 1 049 | 克隆 |
| A:TGGATAGGCTGTTTAATCTCAGATG | Clone | |||
| ZNF32 | S:TGGAAGATATGCCCAGAATGTAG | 60 | 293 | qPCR |
| A:CACAATGGGTACACTCGGG | ||||
| ZNF32 | S:CCGGAATTCCATGTTTGGATTTCCAACAGCTACCC | 58 | 822 | 过表达载体构建 |
| A:CCGCTCGAGACTTACAGGGTGAGCCTTTGTGAGC | Overexpression plasmid construction | |||
| Si-NC | S:UUCUCCGAACGUGUCACGUTT | 60 | 干扰 | |
| A:ACGUGACACGUUCGGAGAATT | Interference | |||
| Si-ZNF32 | S:GGGUAGUCUAACAUUACAUTT | 60 | 干扰 | |
| A:AUGUAAUGUUAGACUACCCTT | Interference | |||
| TBP | S:AACAGCCTCCCACCTTATGC | 60 | 155 | 内参 |
| A:TGCTGCTCCTCCTAGAC | Reference | |||
| UXT | S:GCAAGTGGATTTGGGCTGTAAC | 60 | 180 | 内参 |
| A:ATGGAGTCCTTGGTGAGGTTGT | Reference |
| 分析内容 Analytical content | 分析软件或在线工具 Analytical software or online tools |
|---|---|
| 氨基酸序列比对 Amino acid sequences alignment | DNAMAN |
| 氨基酸序列翻译、同源性比对 Amino acid sequence translation and homology comparison | ORF Finder、Blast(NCBI) |
| 蛋白质理化性质分析Protein physicochemical properties analysis | ExPASy ProtParam |
| 蛋白质磷酸化位点分析 Protein phosphorylation site analysis | NetPhos 3.1 |
| 蛋白质功能结构域分析 Protein functional domain analysis | Conserved Domain |
| 蛋白质二级结构预测 Protein secondary structure prediction | Npsa-prabi |
| 蛋白质三级结构预测 Protein tertiary structure prediction | SWISS-MODEL |
| 蛋白质相互作用预测 Interaction protein prediction | STRING |
| 系统进化树构建 Phylogenetic tree construction | MEGA 5.0 |
表 2 生物信息学分析内容及分析工具
Table 2 Analytical contents and analysis tools for bioinfo-rmatics
| 分析内容 Analytical content | 分析软件或在线工具 Analytical software or online tools |
|---|---|
| 氨基酸序列比对 Amino acid sequences alignment | DNAMAN |
| 氨基酸序列翻译、同源性比对 Amino acid sequence translation and homology comparison | ORF Finder、Blast(NCBI) |
| 蛋白质理化性质分析Protein physicochemical properties analysis | ExPASy ProtParam |
| 蛋白质磷酸化位点分析 Protein phosphorylation site analysis | NetPhos 3.1 |
| 蛋白质功能结构域分析 Protein functional domain analysis | Conserved Domain |
| 蛋白质二级结构预测 Protein secondary structure prediction | Npsa-prabi |
| 蛋白质三级结构预测 Protein tertiary structure prediction | SWISS-MODEL |
| 蛋白质相互作用预测 Interaction protein prediction | STRING |
| 系统进化树构建 Phylogenetic tree construction | MEGA 5.0 |
| 目的基因 Target gene | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 用途 Purpose |
|---|---|---|---|
| P21 | S:AGGGCACGTCTCAGGAGGA A:CAGTCTGCGTTTGGAGTGGTAG | 60 | qPCR |
| P27 | S:CGGCGGTGCCTTTACTT A:GCAGGTCGCTTCCTTATCC | 60 | qPCR |
| P53 | S:CGCCTCAGCACCTTATCCG A:GGCACCACGCACCTCA | 60 | qPCR |
| P57 | S:CAGCCAGAGCATTGGCAATG A:CTGTCCACCTCGGTCCACT | 60 | qPCR |
表3 增殖抑制相关基因引物信息
Table 3 Primers information of genes associated with pro-liferation inhibition
| 目的基因 Target gene | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 用途 Purpose |
|---|---|---|---|
| P21 | S:AGGGCACGTCTCAGGAGGA A:CAGTCTGCGTTTGGAGTGGTAG | 60 | qPCR |
| P27 | S:CGGCGGTGCCTTTACTT A:GCAGGTCGCTTCCTTATCC | 60 | qPCR |
| P53 | S:CGCCTCAGCACCTTATCCG A:GGCACCACGCACCTCA | 60 | qPCR |
| P57 | S:CAGCCAGAGCATTGGCAATG A:CTGTCCACCTCGGTCCACT | 60 | qPCR |
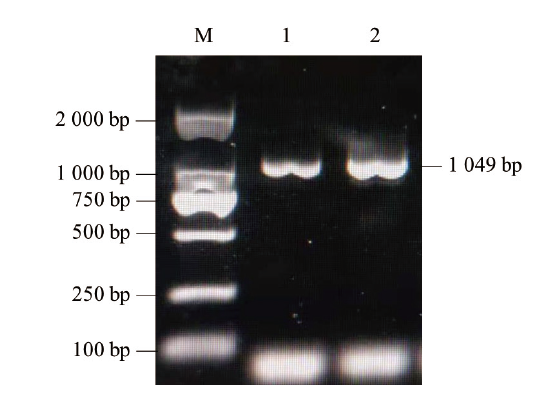
图1 山羊ZNF32基因扩增结果 M:DNA相对分子质量标准DL2000;1-2:山羊ZNF32基因目的条带
Fig. 1 Amplification of ZNF32 gene in goat M:DL2000 DNA marker. 1-2:Target strip of ZNF32 gene in goat
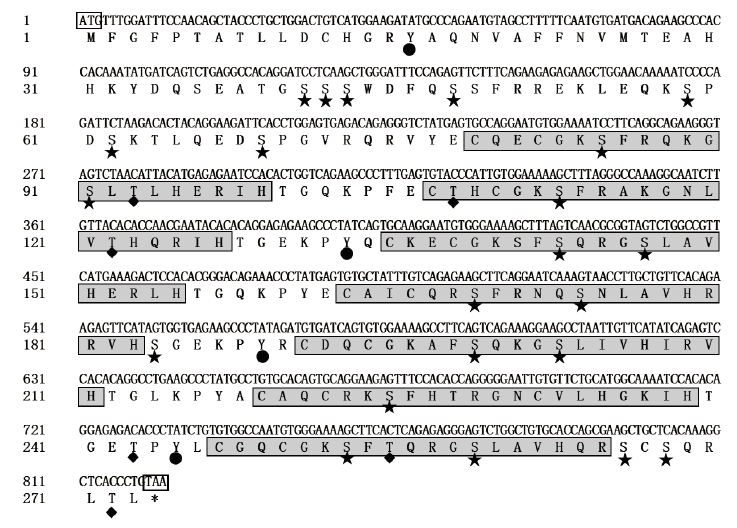
图2 山羊ZNF32基因的ORF序列及推断的氨基酸序列星形:丝氨酸磷酸化位点;四边形:苏氨酸磷酸化位点;圆形:酪氨酸磷酸化位点;阴影框:C2H2锌指结构域;ATG:起始密码子;TAA:终止密码子
Fig. 2 Sequences of ORF and deduced amino acids of ZNF32 gene in goat Stars:Serine phosphorylation sites. Quadrilaterals:Threonine phosphorylation sites. Circles:Tyrosine phosphorylation sites. Shadow boxes:C2H2 zinc finger domains. ATG:The start codon. TAA:The stop codon

图3 山羊ZNF32蛋白疏水性及氨基酸组成分析 A:山羊ZNF32蛋白疏水性分析;B:山羊ZNF32蛋白氨基酸组成分析
Fig. 3 Analysis of hydrophobicity and amino acid compos-ition of ZNF32 protein in goat A:Hydrophobicity analysis of ZNF32 protein in goat. B:Analysis of amino acid composition of ZNF32 protein in goat

图4 山羊ZNF32蛋白质结构及相互作用预测 A:山羊ZNF32蛋白质二级结构预测;B:山羊ZNF32蛋白质三级结构预测;C:ZNF32蛋白质互作网络
Fig. 4 Protein structure and interaction prediction of goat ZNF32 A:Prediction of secondary structure of goat ZNF32 protein. B:Prediction of tertiary structure of goat ZNF32 protein. C:ZNF32 protein interaction network

图5 ZNF32氨基酸序列比对及蛋白系统进化树 A:不同物种ZNF32氨基酸序列比对;B:系统进化树
Fig. 5 Amino acid sequence alignment and protein phylogenetic tree of goat ZNF32 gene A:Alignment of amino acid sequence of ZNF32 in different species. B:Phylogenetic tree

图6 山羊ZNF32组织表达谱 1:背最长肌;2:心;3:肝;4:瘤胃;5:肺;6:臂三头肌;7:股二头肌;8:肾;9:皮下脂肪;10:胰腺;11:小肠;12:腹间脂肪;13:脾;14:大肠。不同大写字母表示差异极显著(P<0.01);不同小写字母表示差异显著(P<0.05);相同字母表示差异不显著(P>0.05).下同
Fig. 6 Expression profile of ZNF32 gene in different tissues of goat 1:Longissimus dorsi;2:heart;3:liver;4:rumen;5:lung;6:triceps brachii;7:biceps femoris;8:kidney;9:subcutaneous;10:pancreas;11:small intestine;12:abdominal fat;13:spleen;14:large intestine. Different capital letters indicate significant differences(P<0.01). Different lowercase letters showed significant differences(P<0.05). The same letters indicate insignificant differences(P>0.05). The same below
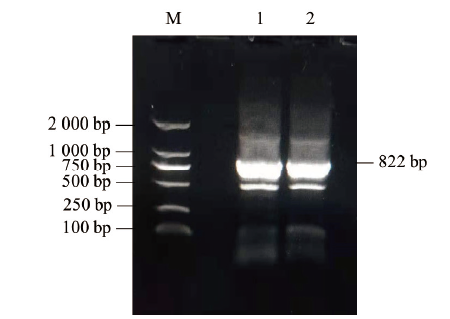
图8 山羊ZNF32基因CDS区扩增结果 M:DNA相对分子质量标准DL2000;1-2:山羊ZNF32基因CDS区扩增结果
Fig. 8 CDS amplification results of goat ZNF32 gene M:DL2000 DNA marker. 1-2:Target strip in the CDS region of goat ZNF32 gene
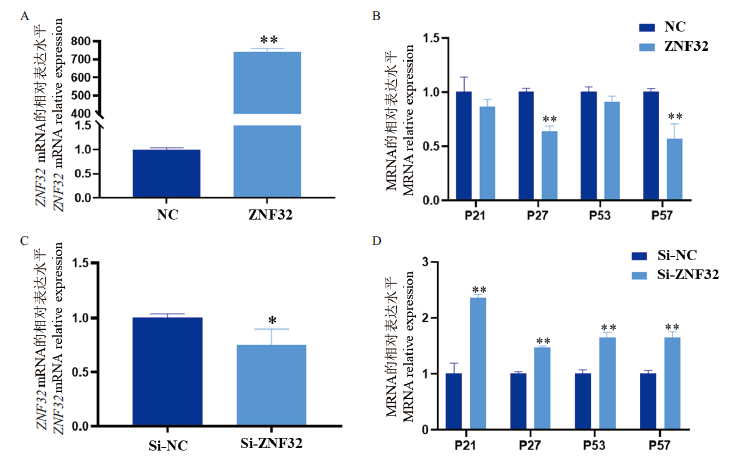
图10 山羊ZNF32基因表达效率及增殖抑制相关基因表达检测 A:过表达ZNF32效率检测;B:过表达ZNF32后增殖抑制相关基因表达检测;C:干扰ZNF32效率检测;D:干扰ZNF32后增殖抑制相关基因检测。*:P<0.05,* *:P<0.01
Fig. 10 Detection of ZNF32 gene expression efficiency and the expressions of genes related to proliferation inhibition in goats A:Efficiency detection of ZNF32 overexpression. B:Expression detection of genes associated with proliferation inhibition after the overexpression of ZNF32. C:Efficiency detection after the interference of ZNF32. D:Detection of genes associated with proliferation inhibition after interference of ZNF32. *:P<0.05,and * *:P<0.01

图11 p53、p21和p27启动子区ZNF32结合序列 A:p53启动子区ZNF32结合序列;B:p21启动子区ZNF32结合序列;C:p27启动子区ZNF32结合序列
Fig. 11 Binding sequence of ZNF32 in p53,p21 and p27 promoter region A:The binding sequence of ZNF32 in p53 promoter region. B:The binding sequence of ZNF32 in p21 promoter region. C:The binding sequence of ZNF32 in p27 promoter region
| [1] |
Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes[J]. Embo J, 1985, 4(6):1609-1614.
doi: 10.1002/j.1460-2075.1985.tb03825.x pmid: 4040853 |
| [2] |
Lee MS, Gippert GP, Soman KV, et al. Three-dimensional solution structure of a single zinc finger DNA-binding domain[J]. Science, 1989, 245(4918):635-637.
pmid: 2503871 |
| [3] | 侯尧, 陈静, 伍春莲. 锌指蛋白32功能的研究进展[J]. 生命的化学, 2020, 40(9):1493-1499. |
| Hou Y, Chen J, Wu CL. Research progress on zinc finger protein 32 function[J]. Chem Life, 2020, 40(9):1493-1499. | |
| [4] |
Han GL, Lu CX, Guo JR, et al. C2H2 zinc finger proteins:master regulators of abiotic stress responses in plants[J]. Front Plant Sci, 2020, 11:115.
doi: 10.3389/fpls.2020.00115 URL |
| [5] | Zarzynska JM. The importance of autophagy regulation in breast cancer development and treatment[J]. Biomed Res Int, 2014, 2014:710345. |
| [6] |
Cannizzaro LA, Aronson MM, Thiesen HJ. Human zinc finger gene ZNF23(Kox16)maps to a zinc finger gene cluster on chromosome 16q22, and ZNF32(Kox30)to chromosome region 10q23-Q24[J]. Hum Genet, 1993, 91(4):383-385.
pmid: 8500793 |
| [7] |
Hong Q, Li C, Xie YS, et al. Kruppel-like factor-15 inhibits the proliferation of mesangial cells[J]. Cell Physiol Biochem, 2012, 29(5/6):893-904.
doi: 10.1159/000178518 URL |
| [8] |
Andreoli V, Gehrau RC, Bocco JL. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death[J]. IUBMB Life, 2010, 62(12):896-905.
doi: 10.1002/iub.396 pmid: 21154818 |
| [9] |
Rokkam P, Gugalavath S, Gift Kumar DK, et al. Prognostic role of hedgehog-GLI1 signaling pathway in aggressive and metastatic breast cancers[J]. Curr Drug Metab, 2020, 21(1):33-43.
doi: 10.2174/1389200221666200122120625 pmid: 31969097 |
| [10] |
Longo M, Raciti GA, Zatterale F, et al. Epigenetic modifications of the Zfp/ZNF423 gene control murine adipogenic commitment and are dysregulated in human hypertrophic obesity[J]. Diabetologia, 2018, 61(2):369-380.
doi: 10.1007/s00125-017-4471-4 pmid: 29067487 |
| [11] |
Matsubara Y, Aoki M, Endo T, et al. Characterization of the expression profiles of adipogenesis-related factors, ZNF423, KLFs and FGF10, during preadipocyte differentiation and abdominal adipose tissue development in chickens[J]. Comp Biochem Physiol B Biochem Mol Biol, 2013, 165(3):189-195.
doi: 10.1016/j.cbpb.2013.04.002 URL |
| [12] |
Chiarella E, Aloisio A, Codispoti B, et al. ZNF521 has an inhibitory effect on the adipogenic differentiation of human adipose-derived mesenchymal stem cells[J]. Stem Cell Rev Rep, 2018, 14(6):901-914.
doi: 10.1007/s12015-018-9830-0 pmid: 29938352 |
| [13] |
Li YY, Gong D, Zhang L, et al. Zinc finger protein 32 promotes breast cancer stem cell-like properties through directly promoting GPER transcription[J]. Cell Death Dis, 2018, 9(12):1162.
doi: 10.1038/s41419-018-1144-2 pmid: 30478301 |
| [14] | Xu Y, Qin Y, Cui JX, et al. microRNA-136-5p regulates gemcitabine resistance in pancreatic cancer via down-regulating ZNF32[J]. Eur Rev Med Pharmacol Sci, 2020, 24(20):10472-10482. |
| [15] |
Wei YY, Li K, Yao SH, et al. Correction:Loss of ZNF32 augments the regeneration of nervous lateral line system through negative regulation of SOX2 transcription[J]. Oncotarget, 2019, 10(67):7179-7180.
doi: 10.18632/oncotarget.27337 URL |
| [16] |
Li YY, Zhang L, Li K, et al. ZNF32 inhibits autophagy through the mTOR pathway and protects MCF-7 cells from stimulus-induced cell death[J]. Sci Rep, 2015, 5:9288.
doi: 10.1038/srep09288 pmid: 25786368 |
| [17] |
Li K, Gao B, Li J, et al. ZNF32 protects against oxidative stress-induced apoptosis by modulating C1QBP transcription[J]. Oncotarget, 2015, 6(35):38107-38126.
doi: 10.18632/oncotarget.5646 pmid: 26497555 |
| [18] |
Li J, Ao J, Li K, et al. ZNF32 contributes to the induction of multidrug resistance by regulating TGF-β receptor 2 signaling in lung adenocarcinoma[J]. Cell Death Dis, 2016, 7(10):e2428.
doi: 10.1038/cddis.2016.328 URL |
| [19] |
Li K, Zhao G, Ao J, et al. ZNF32 induces anoikis resistance through maintaining redox homeostasis and activating Src/FAK signaling in hepatocellular carcinoma[J]. Cancer Lett, 2019, 442:271-278.
doi: S0304-3835(18)30669-4 pmid: 30439540 |
| [20] | 陈宏丽, 杨敬, 尹刚, 等. 锌指蛋白32在口腔鳞状细胞癌中的表达意义及对口腔鳞状细胞癌干细胞的影响[J]. 国际口腔医学杂志, 2019, 46(6):631-639. |
| Chen HL, Yang J, Yin G, et al. Expression of zinc finger protein 32 in oral squamous cell carcinoma and its effect on oral squamous cell carcinoma stem cells[J]. Int J Stomatol, 2019, 46(6):631-639. | |
| [21] |
Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity[J]. N Engl J Med, 2017, 376(3):254-266.
doi: 10.1056/NEJMra1514009 URL |
| [22] |
Blüher M. Obesity:global epidemiology and pathogenesis[J]. Nat Rev Endocrinol, 2019, 15(5):288-298.
doi: 10.1038/s41574-019-0176-8 URL |
| [23] | 池永东, 王永, 胡萌, 等. 山羊不同组织器官的内参基因筛选[J]. 基因组学与应用生物学, 2020, 39(2):561-567. |
| Chi YD, Wang Y, Hu M, et al. Screening of internal reference genes in different tissues and organs of goats[J]. Genom Appl Biol, 2020, 39(2):561-567. | |
| [24] | 林森, 林亚秋, 朱江江, 等. Wnt10b对山羊前体脂肪细胞分化相关基因表达的影响[J]. 畜牧兽医学报, 2018, 49(4):685-692. |
| Lin S, Lin YQ, Zhu JJ, et al. Effect of Wnt10b on the expression of precursor adipocytes differentiation related genes in goat[J]. Chin J Animal Vet Sci, 2018, 49(4):685-692. | |
| [25] | 许晴, 林森, 朱江江, 等. 山羊肌内前体脂肪细胞诱导分化过程中内参基因的表达稳定性分析[J]. 畜牧兽医学报, 2018, 49(5):907-918. |
| Xu Q, Lin S, Zhu JJ, et al. The expression stability analysis of reference genes in the process of goat intramuscular preadipocytes differentiation in goat[J]. Chin J Animal Vet Sci, 2018, 49(5):907-918. | |
| [26] |
Humphrey SJ, James DE, Mann M. Protein phosphorylation:a major switch mechanism for metabolic regulation[J]. Trends Endocrinol Metab, 2015, 26(12):676-687.
doi: 10.1016/j.tem.2015.09.013 URL |
| [27] |
Luo ZJ, Gao X, Lin CQ, et al. Zic2 is an enhancer-binding factor required for embryonic stem cell specification[J]. Mol Cell, 2015, 57(4):685-694.
doi: S1097-2765(15)00008-8 pmid: 25699711 |
| [28] |
Murn J, Zarnack K, Yang YJ, et al. Control of a neuronal morphology program by an RNA-binding zinc finger protein, Unkempt[J]. Genes Dev, 2015, 29(5):501-512.
doi: 10.1101/gad.258483.115 URL |
| [29] | 朱江江, 林亚秋, 王永, 等. 山羊Kruppel样转录因子家族在前体脂肪细胞分化中的表达模式及相关性分析[J]. 中国农业科学, 2019, 52(13):2341-2351. |
| Zhu JJ, Lin YQ, Wang Y, et al. Expression profile and correlations of kruppel like factors during caprine(Capra hircus)preadipocyte differentiation[J]. Sci Agric Sin, 2019, 52(13):2341-2351. | |
| [30] |
Zhao YY, Tang XJ, Huang YH, et al. Interaction of c-Jun and HOTAIR- increased expression of p21 converge in polyphyllin I-inhibited growth of human lung cancer cells[J]. Onco Targets Ther, 2019, 12:10115-10127.
doi: 10.2147/OTT.S226830 URL |
| [31] |
Zhou YY, Wang K, Zhen S, et al. Carfilzomib induces G2/M cell cycle arrest in human endometrial cancer cells via upregulation of p21 Waf1/Cip1 and p27 Kip1[J]. Taiwan J Obstet Gynecol, 2016, 55(6):847-851.
doi: 10.1016/j.tjog.2016.09.003 URL |
| [32] |
Vlachos P, Joseph B. The Cdk inhibitor p57(Kip2)controls LIM-kinase 1 activity and regulates actin cytoskeleton dynamics[J]. Oncogene, 2009, 28(47):4175-4188.
doi: 10.1038/onc.2009.269 pmid: 19734939 |
| [33] |
Kamada R, Toguchi Y, Nomura T, et al. Tetramer formation of tumor suppressor protein p53:Structure, function, and applications[J]. Biopolymers, 2016, 106(4):598-612.
doi: 10.1002/bip.22772 pmid: 26572807 |
| [34] |
Sun CQ, Ma P, Wang YF, et al. KLF15 inhibits cell proliferation in gastric cancer cells via up-regulating CDKN1A/p21 and CDKN1C/p57 expression[J]. Dig Dis Sci, 2017, 62(6):1518-1526.
doi: 10.1007/s10620-017-4558-2 URL |
| [35] |
Wang ZF, Yu CZ, Wang H. HOXA5 inhibits the proliferation and induces the apoptosis of cervical cancer cells via regulation of protein kinase B and p27[J]. Oncol Rep, 2019, 41(2):1122-1130.
doi: 10.3892/or.2018.6874 pmid: 30483748 |
| [36] |
Yang YP, Qin RH, Zhao JJ, et al. BOP1 silencing suppresses gastric cancer proliferation through p53 modulation[J]. Curr Med Sci, 2021, 41(2):287-296.
doi: 10.1007/s11596-021-2345-y URL |
| [37] | 孙凯, 王伟, 雷尚通, 等. microRNA-221通过抑制CDKN1C/p57表达促进结肠癌细胞增殖[J]. 南方医科大学学报, 2011, 31(11):1885-1889. |
| Sun K, Wang W, Lei ST, et al. microRNA-221 promotes colon carcinoma cell proliferation in vitro by inhibiting CDKN1C/p57 expression[J]. J South Med Univ, 2011, 31(11):1885-1889. |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 李英, 岳祥华. DNA甲基化在解析毛竹自然变异中的应用[J]. 生物技术通报, 2023, 39(7): 48-55. |
| [5] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [6] | 马钰静, 段春辉, 贺名扬, 张英杰, 杨若晨, 王泳, 刘月琴. 敲除G0S2基因对绵羊卵巢颗粒细胞增殖、类固醇激素及相关基因表达的影响[J]. 生物技术通报, 2023, 39(6): 325-334. |
| [7] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [8] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [9] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [10] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [11] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [12] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [13] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [14] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [15] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||