








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 29-40.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0520
马赛买1( ), 李同源1, 马燕军1, 韩富军2, 彭海2, 孔维宝1(
), 李同源1, 马燕军1, 韩富军2, 彭海2, 孔维宝1( )
)
收稿日期:2023-06-02
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
孔维宝,男,博士,教授,研究方向:微生物资源与技术;E-mail: kongwb@nwnu.edu.cn作者简介:马赛买,女,硕士研究生,研究方向:微生物技术;E-mail: silmah79@163.com
基金资助:
MA Sai-mai1( ), LI Tong-yuan1, MA Yan-jun1, HAN Fu-jun2, PENG Hai2, KONG Wei-bao1(
), LI Tong-yuan1, MA Yan-jun1, HAN Fu-jun2, PENG Hai2, KONG Wei-bao1( )
)
Received:2023-06-02
Published:2023-10-26
Online:2023-11-28
摘要:
病虫害严重影响农作物的品质、产量和安全。生物防治作为一种绿色、安全、有效防控病虫害的技术方法越来越受到人们关注。几丁质酶(EC 3.2.1.14)是一类广泛存在于微生物和植物中的糖苷水解酶,可有效降解病原真菌细胞壁、昆虫及线虫体壁中的几丁质,抑制病原真菌孢子萌发和菌丝生长,以及昆虫和线虫的发育,在农作物病虫害生物防治中具有重要作用和广泛的应用前景。本文综述了几丁质酶的种类和来源、生防机理,以及基于几丁质酶的转基因技术在作物病虫害防治中的应用等方面的进展,以期为几丁质酶的进一步研究和应用提供信息参考。
马赛买, 李同源, 马燕军, 韩富军, 彭海, 孔维宝. 几丁质酶在农作物病虫害生物防治中的研究进展[J]. 生物技术通报, 2023, 39(10): 29-40.
MA Sai-mai, LI Tong-yuan, MA Yan-jun, HAN Fu-jun, PENG Hai, KONG Wei-bao. Research Progress in Chitinase Involving in the Biocontrol of Crop Diseases and Pests[J]. Biotechnology Bulletin, 2023, 39(10): 29-40.

图1 黏质沙雷氏菌18家族几丁质酶的结构概述图中显示了ChiA、ChiB的晶体结构和ChiC的催化结构域,以及ChiC中FnIII和CBM5/ 12结构域同系物的NMR结构;螺旋为深蓝色,β股为青色;催化酸(Glu)的侧链显示为黄色
Fig. 1 Structural overview of the family 18 chitinases of S. marcescens The picture shows the crystal structures of ChiA, ChiB and the catalytic domain of ChiC, and the NMR structures of homologues of the FnⅢ and the CBM5/12 domain in ChiC. Helices are colored dark blue, β-strands are colored cyan. The side chain of the catalytic acid(a Glu)is shown in yellow
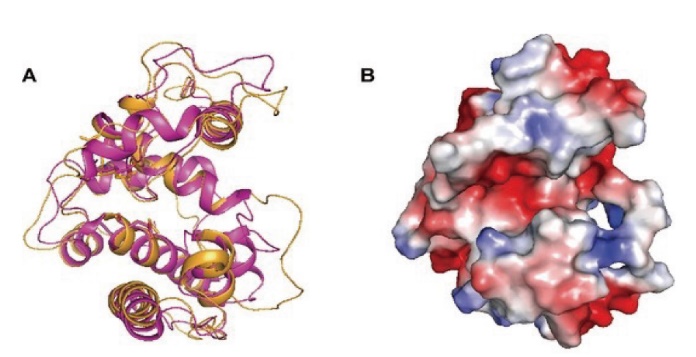
图2 NbchiA的三维模型 A:NbchiA模型的整体折叠(橙色)和NbchiA与S. coelicor几丁质酶单体(紫色)的叠加;B:NbchiA模型的分子表面,所有与催化和结合相关的残基都位于一个口袋中
Fig. 2 Three-dimensional model of NbchiA A: Cartoon representation of the overall fold of NbchiA model(orange)and superimposition of NbchiA and S. coelicolor chitinase(purple)monomers. B: Molecular surface of the NbchiA model, all of the residues related to catalysis and binding are located in a pocket
| [1] |
Fernandez MR, Abdellatif L, Lokuruge P, et al. Root disease and fungal populations in organic crops under different tillage-cropping systems[J]. Crop Sci, 2022, 62(3): 1288-1304.
doi: 10.1002/csc2.v62.3 URL |
| [2] |
Leybourne DJ, Storer KE, Berry P, et al. Development of a pest threshold decision support system for minimising damage to winter wheat from wheat bulb fly, Delia coarctata[J]. Ann Appl Biol, 2022, 180(1): 118-131.
doi: 10.1111/aab.v180.1 URL |
| [3] |
Rahman M, Islam T, Jett L, et al. Biocontrol agent, biofumigation, and grafting with resistant rootstock suppress soil-borne disease and improve yield of tomato in West Virginia[J]. Crop Prot, 2021, 145: 105630.
doi: 10.1016/j.cropro.2021.105630 URL |
| [4] |
Withers AJ, Rice A, de Boer J, et al. The distribution of covert microbial natural enemies of a globally invasive crop pest, fall armyworm, in Africa: enemy release and spillover events[J]. J Anim Ecol, 2022, 91(9): 1826-1841.
doi: 10.1111/jane.v91.9 URL |
| [5] | Iftikhar Y, Mubeen M, Sajid A, et al. Effects of tomato leaf curl virus on growth and yield parameters of tomato crop[J]. Arab J Plant Prot, 2021, 39(1): 79-83. |
| [6] |
Mazumdar P, Singh P, Kethiravan D, et al. Late blight in tomato: insights into the pathogenesis of the aggressive pathogen Phytophthora infestans and future research priorities[J]. Planta, 2021, 253(6): 119.
doi: 10.1007/s00425-021-03636-x pmid: 33963935 |
| [7] |
Li J, Zou CG, Xu JP, et al. Molecular mechanisms of nematode-nematophagous microbe interactions: basis for biological control of plant-parasitic nematodes[J]. Annu Rev Phytopathol, 2015, 53: 67-95.
doi: 10.1146/annurev-phyto-080614-120336 pmid: 25938277 |
| [8] |
Kenney E, Eleftherianos I. Entomopathogenic and plant pathogenic nematodes as opposing forces in agriculture[J]. Int J Parasitol, 2016, 46(1): 13-19.
doi: 10.1016/j.ijpara.2015.09.005 pmid: 26527129 |
| [9] |
Park MH, Walpola BC, Kim SJ, et al. Control effect of root-knot nematode(Meloidogyne incognita)by biological nematicide[J]. Korean J Soil Sci Fertil, 2012, 45(2): 162-168.
doi: 10.7745/KJSSF.2012.45.2.162 URL |
| [10] |
Zitnick-Anderson K, del Río Mendoza LE, Forster S, et al. Associations among the communities of soil-borne pathogens, soil edaphic properties and disease incidence in the field pea root rot complex[J]. Plant Soil, 2020, 457(1): 339-354.
doi: 10.1007/s11104-020-04745-4 |
| [11] |
Ren J, Tong J, Li PH, et al. Chitosan is an effective inhibitor against potato dry rot caused by Fusarium oxysporum[J]. Physiol Mol Plant Pathol, 2021, 113: 101601.
doi: 10.1016/j.pmpp.2021.101601 URL |
| [12] |
Viswanathan R, Selvakumar R, Manivannan K, et al. Behaviour of soil borne inoculum of Colletotrichum falcatum in causing red rot in sugarcane varieties with varying disease resistance[J]. Sugar Tech, 2020, 22(3): 485-497.
doi: 10.1007/s12355-020-00800-7 |
| [13] |
Darwesh OM, Elshahawy IE. Silver nanoparticles inactivate sclerotial formation in controlling white rot disease in onion and garlic caused by the soil borne fungus Stromatinia cepivora[J]. Eur J Plant Pathol, 2021, 160(4): 917-934.
doi: 10.1007/s10658-021-02296-7 |
| [14] | Singh G, Bhalla A, Bhatti JS, et al. Potential of chitinases as a biopesticide against agriculturally harmful fungi and insects[J]. Res Rev J Microbiol Biotechnol, 2014, 3: 27-32. |
| [15] | Kaur H, Garg H. Pesticides: environmental impacts and management strategies[M]//Soloneski S, Ed. Pesticides - Toxic Aspects. 2014:187-230. |
| [16] |
Berini F, Casartelli M, Montali A, et al. Metagenome-sourced microbial chitinases as potential insecticide proteins[J]. Front Microbiol, 2019, 10: 1358.
doi: 10.3389/fmicb.2019.01358 pmid: 31275279 |
| [17] |
Rojas EC, Jensen B, Jørgensen HJL, et al. Selection of fungal endophytes with biocontrol potential against Fusarium head blight in wheat[J]. Biol Contr, 2020, 144: 104222.
doi: 10.1016/j.biocontrol.2020.104222 URL |
| [18] |
Barratt BIP, Moran VC, Bigler F, et al. The status of biological control and recommendations for improving uptake for the future[J]. BioControl, 2018, 63(1): 155-167.
doi: 10.1007/s10526-017-9831-y URL |
| [19] |
Elnahal ASM, El-Saadony MT, Saad AM, et al. The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: a review[J]. Eur J Plant Pathol, 2022, 162(4): 759-792.
doi: 10.1007/s10658-021-02393-7 |
| [20] |
Katembo N, Witkowski ETF, Simelane DO, et al. Impact of biocontrol agents on Lantana camara in an inland area of South Africa[J]. BioControl, 2020, 65(2): 143-154.
doi: 10.1007/s10526-019-09991-9 |
| [21] |
Köhl J, Booij K, Kolnaar R, et al. Ecological arguments to reconsider data requirements regarding the environmental fate of microbial biocontrol agents in the registration procedure in the European Union[J]. BioControl, 2019, 64(5): 469-487.
doi: 10.1007/s10526-019-09964-y |
| [22] |
Macena AMF, Kobori NN, Mascarin GM, et al. Antagonism of Trichoderma-based biofungicides against Brazilian and North American isolates of Sclerotinia sclerotiorum and growth promotion of soybean[J]. BioControl, 2020, 65(2): 235-246.
doi: 10.1007/s10526-019-09976-8 |
| [23] |
Vasconcelos S, Jonsson M, Heleno R, et al. A meta-analysis of biocontrol potential and herbivore pressure in olive crops: does integrated pest management make a difference?[J]. Basic Appl Ecol, 2022, 63: 115-124.
doi: 10.1016/j.baae.2022.05.009 URL |
| [24] |
Zhu WX, Duan YW, Chen JQ, et al. SERCA interacts with chitin synthase and participates in cuticular chitin biogenesis in Drosophila[J]. Insect Biochem Mol Biol, 2022, 145: 103783.
doi: 10.1016/j.ibmb.2022.103783 URL |
| [25] |
Grover A. Plant chitinases: genetic diversity and physiological roles[J]. Crit Rev Plant Sci, 2012, 31(1): 57-73.
doi: 10.1080/07352689.2011.616043 URL |
| [26] |
Berini F, Caccia S, Franzetti E, et al. Effects of Trichoderma viride chitinases on the peritrophic matrix of Lepidoptera[J]. Pest Manag Sci, 2016, 72(5): 980-989.
doi: 10.1002/ps.2016.72.issue-5 URL |
| [27] |
Berini F, Presti I, Beltrametti F, et al. Production and characterization of a novel antifungal chitinase identified by functional screening of a suppressive-soil metagenome[J]. Microb Cell Fact, 2017, 16(1): 16.
doi: 10.1186/s12934-017-0634-8 pmid: 28137256 |
| [28] |
Hjort K, Presti I, Elväng A, et al. Bacterial chitinase with phytopathogen control capacity from suppressive soil revealed by functional metagenomics[J]. Appl Microbiol Biotechnol, 2014, 98(6): 2819-2828.
pmid: 24121932 |
| [29] |
Adrangi S, Faramarzi MA. From bacteria to human: a journey into the world of chitinases[J]. Biotechnol Adv, 2013, 31(8): 1786-1795.
doi: 10.1016/j.biotechadv.2013.09.012 pmid: 24095741 |
| [30] |
Huang QS, Xie XL, Liang G, et al. The GH18 family of chitinases: their domain architectures, functions and evolutions[J]. Glycobiology, 2012, 22(1): 23-34.
doi: 10.1093/glycob/cwr092 URL |
| [31] |
Li H, Greene LH. Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding[J]. PLoS One, 2010, 5(1): e8654.
doi: 10.1371/journal.pone.0008654 URL |
| [32] | Udaya Prakash NA, Jayanthi M, Sabarinathan R, et al. Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases[J]. J Mol Evol, 2010, 70 |
| 5):466-478. | |
| [33] |
Cao SL, Wang Y, Li ZQ, et al. Genome-wide identification and expression analyses of the chitinases under cold and osmotic stress in Ammopiptanthus nanus[J]. Genes, 2019, 10(6): 472.
doi: 10.3390/genes10060472 URL |
| [34] |
Chen JJ, Piao YL, Liu YM, et al. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance[J]. Plant Sci, 2018, 270: 257-267.
doi: 10.1016/j.plantsci.2018.02.017 URL |
| [35] |
Bordoloi KS, Krishnatreya DB, Baruah PM, et al. Genome-wide identification and expression profiling of chitinase genes in tea(Camellia sinensis(L.) O. Kuntze)under biotic stress conditions[J]. Physiol Mol Biol Plants, 2021, 27(2): 369-385.
doi: 10.1007/s12298-021-00947-x |
| [36] |
Cretoiu MS, Kielak AM, Abu Al-Soud W, et al. Mining of unexplored habitats for novel chitinases—chiA as a helper gene proxy in metagenomics[J]. Appl Microbiol Biotechnol, 2012, 94(5): 1347-1358.
doi: 10.1007/s00253-012-4057-5 URL |
| [37] |
Metcalfe AC, Krsek M, Gooday GW, et al. Molecular analysis of a bacterial chitinolytic community in an upland pasture[J]. Appl Environ Microbiol, 2002, 68(10): 5042-5050.
doi: 10.1128/AEM.68.10.5042-5050.2002 URL |
| [38] |
Vaaje-Kolstad G, Horn SJ, Sørlie M, et al. The chitinolytic machinery of Serratia marcescens —a model system for enzymatic degradation of recalcitrant polysaccharides[J]. FEBS J, 2013, 280(13): 3028-3049.
doi: 10.1111/febs.12181 pmid: 23398882 |
| [39] |
Dahiya D, Pilli A, Chirra PRR, et al. Morphological and structural characterization of chitin as a substrate for the screening, production, and molecular characterization of chitinase by Bacillus velezensis[J]. Environ Sci Pollut Res Int, 2022, 29(57): 86550-86561.
doi: 10.1007/s11356-022-22166-x |
| [40] |
Jankiewicz U, Brzezinska MS, Saks E. Identification and characterization of a chitinase of Stenotrophomonas maltophilia, a bacterium that is antagonistic towards fungal phytopathogens[J]. J Biosci Bioeng, 2012, 113(1): 30-35.
doi: 10.1016/j.jbiosc.2011.08.023 pmid: 21945415 |
| [41] |
García-Fraga B, da Silva AF, López-Seijas J, et al. Functional expression and characterization of a chitinase from the marine archaeon Halobacterium salinarum CECT 395 in Escherichia coli[J]. Appl Microbiol Biotechnol, 2014, 98(5): 2133-2143.
doi: 10.1007/s00253-013-5124-2 pmid: 23893326 |
| [42] |
Tuveng TR, Hagen LH, Mekasha S, et al. Genomic, proteomic and biochemical analysis of the chitinolytic machinery of Serratia marcescens BJL200[J]. Biochim Biophys Acta Proteins Proteom, 2017, 1865(4): 414-421.
doi: 10.1016/j.bbapap.2017.01.007 URL |
| [43] |
Mehmood MA, Gai YB, Zhuang QC, et al. Aeromonas caviae CB101 contains four chitinases encoded by a single gene Chi1[J]. Mol Biotechnol, 2010, 44(3): 213-220.
doi: 10.1007/s12033-009-9227-z URL |
| [44] |
Zhang WJ, Ma JW, Yan QJ, et al. Biochemical characterization of a novel acidic chitinase with antifungal activity from Paenibacillus xylanexedens Z2-4[J]. Int J Biol Macromol, 2021, 182: 1528-1536.
doi: 10.1016/j.ijbiomac.2021.05.111 URL |
| [45] |
Han B, Zhou K, Li ZH, et al. Characterization of the first fungal glycosyl hydrolase family 19 chitinase(NbchiA)from Nosema bombycis(Nb)[J]. J Eukaryot Microbiol, 2016, 63(1): 37-45.
doi: 10.1111/jeu.2016.63.issue-1 URL |
| [46] |
Hartl L, Zach S, Seidl-Seiboth V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential[J]. Appl Microbiol Biotechnol, 2012, 93(2): 533-543.
doi: 10.1007/s00253-011-3723-3 pmid: 22134638 |
| [47] | Seidl-Seiboth V, Ihrmark K, Druzhinina I, et al. Molecular evolution of Trichoderma chitinases[M]//Biotechnology and Biology of Trichoderma. Amsterdam: Elsevier, 2014: 67-78. |
| [48] |
Gruber S, Seidl-Seiboth V. Self versus non-self: fungal cell wall degradation in Trichoderma[J]. Microbiology, 2012, 158(Pt 1): 26-34.
doi: 10.1099/mic.0.052613-0 URL |
| [49] |
Salvador R, Ferrelli ML, Sciocco-Cap A, et al. Analysis of a chitinase from EpapGV, a fast killing betabaculovirus[J]. Virus Genes, 2014, 48(2): 406-409.
doi: 10.1007/s11262-013-1019-7 pmid: 24297310 |
| [50] |
Oh S, Kim DH, Patnaik BB, et al. Molecular and immunohistochemical characterization of the chitinase gene from Pieris rapae granulovirus[J]. Arch Virol, 2013, 158(8): 1701-1718.
doi: 10.1007/s00705-013-1649-z URL |
| [51] | Abulikemu S, Yesilyurt A, Gencer D, et al. Comparison of the potential activities of viral and bacterial chitinases[J]. Egypt J Biol Pest Contr, 2021, 31(1):1-7. |
| [52] |
Wang QH, Qu LJ, Zhang ZL, et al. Characterization of a novel chitinase, DkChi, from Dendrolimus kikuchii nucleopolyhedrovirus[J]. Arch Virol, 2013, 158(12): 2523-2530.
doi: 10.1007/s00705-013-1775-7 URL |
| [53] |
Di Maro A, Terracciano I, Sticco L, et al. Purification and characterization of a viral chitinase active against plant pathogens and herbivores from transgenic tobacco[J]. J Biotechnol, 2010, 147(1): 1-6.
doi: 10.1016/j.jbiotec.2010.03.005 pmid: 20302895 |
| [54] |
Parmar N, Singh KH, Sharma D, et al. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review[J]. 3 Biotech, 2017, 7(4): 239.
doi: 10.1007/s13205-017-0870-y pmid: 28702937 |
| [55] |
Malik A, Preety. Purification and properties of plant chitinases: a review[J]. J Food Biochem, 2019, 43(3): e12762.
doi: 10.1111/jfbc.2019.43.issue-3 URL |
| [56] |
Toufiq N, Tabassum B, Bhatti MU, et al. Improved antifungal activity of barley derived chitinase I gene that overexpress a 32kDa recombinant chitinase in Escherichia coli host[J]. Braz J Microbiol, 2018, 49(2): 414-421.
doi: S1517-8382(16)31095-4 pmid: 29146152 |
| [57] |
Tariq M, Khan A, Tabassum B, et al. Antifungal activity of chitinase II against Colletotrichum falcatum Went. causing red rot disease in transgenic sugarcane[J]. Turk J Biol, 2018, 42(1): 45-53.
doi: 10.3906/biy-1709-17 URL |
| [58] |
Garg N, Gupta H. Isolation and purification of fungal pathogen(Macrophomina phaseolina)induced chitinase from moth beans(Phaseolus aconitifolius)[J]. J Pharm Bioallied Sci, 2010, 2(1): 38-43.
doi: 10.4103/0975-7406.62708 URL |
| [59] |
Kabir SR, Rahman MM, Tasnim S, et al. Purification and characterization of a novel chitinase from Trichosanthes dioica seed with antifungal activity[J]. Int J Biol Macromol, 2016, 84: 62-68.
doi: 10.1016/j.ijbiomac.2015.12.006 URL |
| [60] |
Jacquiod S, Franqueville L, Cécillon S, et al. Soil bacterial community shifts after chitin enrichment: an integrative metagenomic approach[J]. PLoS One, 2013, 8(11): e79699.
doi: 10.1371/journal.pone.0079699 URL |
| [61] |
Cretoiu MS, Korthals GW, Visser JHM, et al. Chitin amendment increases soil suppressiveness toward plant pathogens and modulates the actinobacterial and oxalobacteraceal communities in an experimental agricultural field[J]. Appl Environ Microbiol, 2013, 79(17): 5291-5301.
doi: 10.1128/AEM.01361-13 URL |
| [62] | Berini F, Casciello C, Marcone GL, et al. Metagenomics: novel enzymes from non-culturable microbes[J]. FEMS Microbiol Lett, 2017, 364(21): fnx211. |
| [63] |
Ekkers DM, Cretoiu MS, Kielak AM, et al. The great screen anomaly—a new frontier in product discovery through functional metagenomics[J]. Appl Microbiol Biotechnol, 2012, 93(3): 1005-1020.
doi: 10.1007/s00253-011-3804-3 URL |
| [64] |
Dai YM, Yang F, Liu X, et al. The discovery and characterization of a novel chitinase with dual catalytic domains from a Qinghai-Tibetan Plateau wetland soil metagenome[J]. Int J Biol Macromol, 2021, 188: 482-490.
doi: 10.1016/j.ijbiomac.2021.07.153 URL |
| [65] |
Li RK, Hu YJ, Ng TB, et al. Expression and biochemical characterization of a novel chitinase ChiT-7 from the metagenome in the soil of a mangrove tidal flat in China[J]. Int J Biol Macromol, 2020, 158: 1125-1134.
doi: S0141-8130(20)33113-5 pmid: 32360969 |
| [66] |
Berini F, Katz C, Gruzdev N, et al. Microbial and viral chitinases: attractive biopesticides for integrated pest management[J]. Biotechnol Adv, 2018, 36(3): 818-838.
doi: S0734-9750(18)30002-8 pmid: 29305895 |
| [67] |
Swiontek Brzezinska M, Jankiewicz U, Burkowska A, et al. Chitinolytic microorganisms and their possible application in environmental protection[J]. Curr Microbiol, 2014, 68(1): 71-81.
doi: 10.1007/s00284-013-0440-4 pmid: 23989799 |
| [68] |
Yan RX, Hou JH, Ding DF, et al. In vitro antifungal activity and mechanism of action of chitinase against four plant pathogenic fungi[J]. J Basic Microbiol, 2008, 48(4): 293-301.
doi: 10.1002/jobm.v48:4 URL |
| [69] |
Moka SM, Singh N, Buttar DS. Identification of potential native chitinase-producing Trichoderma spp. and its efficacy against damping-off in onion[J]. Eur J Plant Pathol, 2021, 161(2): 289-300.
doi: 10.1007/s10658-021-02321-9 |
| [70] |
Asif T, Javed U, Zafar SB, et al. Bioconversion of colloidal chitin using novel chitinase from Glutamicibacter uratoxydans exhibiting anti-fungal potential by hydrolyzing chitin within fungal cell wall[J]. Waste Biomass Valorization, 2020, 11(8): 4129-4143.
doi: 10.1007/s12649-019-00746-2 |
| [71] |
Akram F, Haq IU, Roohi A, et al. Acinetobacter indicus CCS-12: a new bacterial source for the production and biochemical characterization of thermostable chitinase with promising antifungal activity[J]. Waste Biomass Valorization, 2022, 13(7): 3371-3388.
doi: 10.1007/s12649-022-01753-6 |
| [72] | Sarker KN, Das Mohapatra PK, Dutta S. Use of low cost natural resources for enhanced chitinase production and optimization using CCD and RSM: a new initiative for bio-control of plant pathogen[J]. Indian Phytopathol, 2019, 72(2): 281-300. |
| [73] |
Kramer KJ, Muthukrishnan S. Insect chitinases: molecular biology and potential use as biopesticides[J]. Insect Biochem Mol Biol, 1997, 27(11): 887-900.
doi: 10.1016/S0965-1748(97)00078-7 URL |
| [74] |
Tu S, Qiu XH, Cao L, et al. Expression and characterization of the chitinases from Serratia marcescens GEI strain for the control of Varroa destructor, a honey bee parasite[J]. J Invertebr Pathol, 2010, 104(2): 75-82.
doi: 10.1016/j.jip.2010.02.002 URL |
| [75] |
Zhong WF, Ding SJ, Guo HF. The chitinase C gene PsChiC from Pseudomonas sp. and its synergistic effects on larvicidal activity[J]. Genet Mol Biol, 2015, 38(3): 366-372.
doi: 10.1590/S1415-475738320140320 URL |
| [76] |
Suganthi M, Senthilkumar P, Arvinth S, et al. Chitinase from Pseudomonas fluorescens and its insecticidal activity against Helopeltis theivora[J]. J Gen Appl Microbiol, 2017, 63(4): 222-227.
doi: 10.2323/jgam.2016.11.001 pmid: 28680004 |
| [77] |
Singh AK, Singh A, Joshi P. Combined application of chitinolytic bacterium Paenibacillus sp. D1 with low doses of chemical pesticides for better control of Helicoverpa armigera[J]. Int J Pest Manag, 2016, 62(3): 222-227.
doi: 10.1080/09670874.2016.1167267 URL |
| [78] | Aggarwal C, Paul S, Tripathi V, et al. Chitinase producing Serratia marcescens for biocontrol of Spodoptera litura(Fab)and studies on its chitinolytic activities[J]. Annals of Agricultural Research, 2015, 36(2):132-137. |
| [79] |
Chandrasekaran R, Revathi K, Nisha S, et al. Physiological effect of chitinase purified from Bacillus subtilis against the tobacco cutworm Spodoptera litura Fab[J]. Pestic Biochem Physiol, 2012, 104(1): 65-71.
doi: 10.1016/j.pestbp.2012.07.002 URL |
| [80] |
Danişmazoğlu M, Demİr İ, Sezen K, et al. Cloning and expression of chitinase A, B, and C(chiA, chiB, chiC)genes from Serratia marcescens originating from Helicoverpa armigera and determining their activities[J]. Turk J Biol, 2015, 39: 78-87.
doi: 10.3906/biy-1404-31 URL |
| [81] |
Rathinam M, Marimuthu SK, Tyagi S, et al. Characterization and in planta validation of a CHI4 chitinase from Cajanus platycarpus(Benth.) Maesen for its efficacy against pod borer, Helicoverpa armigera(Hübner)[J]. Pest Manag Sci, 2021, 77(5): 2337-2349.
doi: 10.1002/ps.6260 pmid: 33421295 |
| [82] |
Hu DQ, Luo SH, Abudunasier M, et al. The effect of group IV chitinase, HaCHT4, on the chitin content of the peritrophic matrix(PM)during larval growth and development of Helicoverpa armigera[J]. Pest Manag Sci, 2022, 78(5): 1815-1823.
doi: 10.1002/ps.v78.5 URL |
| [83] |
Ahmad G, Khan A, Khan AA, et al. Biological control: a novel strategy for the control of the plant parasitic nematodes[J]. Antonie Van Leeuwenhoek, 2021, 114(7): 885-912.
doi: 10.1007/s10482-021-01577-9 |
| [84] |
Page AP, Stepek G, Winter AD, et al. Enzymology of the nematode cuticle: a potential drug target?[J]. Int J Parasitol Drugs Drug Resist, 2014, 4(2): 133-141.
doi: 10.1016/j.ijpddr.2014.05.003 URL |
| [85] |
Castaneda-Alvarez C, Prodan S, Rosales IM, et al. Exoenzymes and metabolites related to the nematicidal effect of rhizobacteria on Xiphinema index Thorne & Allen[J]. J Appl Microbiol, 2016, 120(2): 413-424.
doi: 10.1111/jam.12987 pmid: 26541369 |
| [86] |
Chan YL, Cai D, Taylor PWJ, et al. Adverse effect of the chitinolytic enzyme PjCHI-1 in transgenic tomato on egg mass production and embryonic development of Meloidogyne incognita[J]. Plant Pathol, 2010, 59(5): 922-930.
doi: 10.1111/ppa.2010.59.issue-5 URL |
| [87] |
Wei LH, Shao Y, Wan JW, et al. Isolation and characterization of a rhizobacterial antagonist of root-knot nematodes[J]. PLoS One, 2014, 9(1): e85988.
doi: 10.1371/journal.pone.0085988 URL |
| [88] |
Lee YS, Kim KY. Statistical optimization of medium components for chitinase production byPseudomonas fluorescensstrain HN1205: role of chitinase on egg hatching inhibition of root-knot nematode[J]. Biotechnol Biotechnol Equip, 2015, 29(3): 470-478.
doi: 10.1080/13102818.2015.1010702 URL |
| [89] |
Lee YS, Nguyen XH, Naing KW, et al. Role of lytic enzymes secreted by Lysobacter capsici YS1215 in the control of root-knot nematode of tomato plants[J]. Indian J Microbiol, 2015, 55(1): 74-80.
doi: 10.1007/s12088-014-0499-z URL |
| [90] |
Cletus J, Balasubramanian V, Vashisht D, et al. Transgenic expression of plant chitinases to enhance disease resistance[J]. Biotechnol Lett, 2013, 35(11): 1719-1732.
doi: 10.1007/s10529-013-1269-4 pmid: 23794096 |
| [91] |
Ntui VO, Azadi P, Thirukkumaran G, et al. Increased resistance to fusarium wilt in transgenic tobacco lines co-expressing chitinase and wasabi defensin genes[J]. Plant Pathol, 2011, 60(2): 221-231.
doi: 10.1111/ppa.2011.60.issue-2 URL |
| [92] |
Zarinpanjeh N, Motallebi M, Zamani MR, et al. Enhanced resistance to Sclerotinia sclerotiorum in Brassica napus by co-expression of defensin and chimeric chitinase genes[J]. J Appl Genet, 2016, 57(4): 417-425.
pmid: 26862081 |
| [93] |
Amian AA, Papenbrock J, Jacobsen HJ, et al. Enhancing transgenic pea(Pisum sativum L.)resistance against fungal diseases through stacking of two antifungal genes(chitinase and glucanase)[J]. GM Crops, 2011, 2(2): 104-109.
doi: 10.4161/gmcr.2.2.16125 pmid: 21971070 |
| [94] |
Ahmad Mir Z, Ali S, Singh A, et al. In silico analysis and overexpression of chitinase class IV gene in Brassica juncea improves resistance against Alternaria brassicae[J]. Ind Crops Prod, 2021, 169: 113555.
doi: 10.1016/j.indcrop.2021.113555 URL |
| [95] |
Wang F, Yang S, Wang YS, et al. Overexpression of Chitinase gene enhances resistance to Colletotrichum gloeosporioides and Alternaria alternata in apple(Malus × domestica)[J]. Sci Hortic, 2021, 277: 109779.
doi: 10.1016/j.scienta.2020.109779 URL |
| [96] |
Zhang XJ, Huang YJ, Harvey PR, et al. Enhancing plant disease suppression by Burkholderia vietnamiensis through chromosomal integration of Bacillus subtilis chitinase gene Chi113[J]. Biotechnol Lett, 2012, 34(2): 287-293.
doi: 10.1007/s10529-011-0760-z URL |
| [97] |
Navarro-González SS, Ramírez-Trujillo JA, Peña-Chora G, et al. Enhanced tolerance against a fungal pathogen and insect resistance in transgenic tobacco plants overexpressing an endochitinase gene from Serratia marcescens[J]. Int J Mol Sci, 2019, 20(14): 3482.
doi: 10.3390/ijms20143482 URL |
| [1] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [2] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [3] | 章乐乐, 王冠, 柳凤, 胡汉桥, 任磊. 芒果炭疽病拮抗菌分离、鉴定及生防机制研究[J]. 生物技术通报, 2023, 39(4): 277-287. |
| [4] | 易希, 廖红东, 郑井元. 植物内生真菌防治根结线虫研究进展[J]. 生物技术通报, 2023, 39(3): 43-51. |
| [5] | 王伟宸, 赵进, 黄薇颐, 郭芯竹, 李婉颖, 张卓. 芽胞杆菌代谢产物防治三种常见植物病原真菌的研究进展[J]. 生物技术通报, 2023, 39(3): 59-68. |
| [6] | 杨东亚, 祁瑞雪, 李昭轩, 林薇, 马慧, 张雪艳. 黄瓜茄病镰刀菌拮抗芽孢杆菌的筛选、鉴定及促生效果[J]. 生物技术通报, 2023, 39(2): 211-220. |
| [7] | 罗宁, 焦阳, 茆振川, 李惠霞, 谢丙炎. 木霉菌对根结线虫和孢囊线虫防治机理研究进展[J]. 生物技术通报, 2023, 39(2): 35-50. |
| [8] | 林鹰, 杨文莉, 周玲艳, 姜大刚. 农业转基因核酸标准物质研究进展[J]. 生物技术通报, 2022, 38(8): 52-59. |
| [9] | 祖雪, 周瑚, 朱华珺, 任佐华, 刘二明. 枯草芽孢杆菌K-268的分离鉴定及对水稻稻瘟病的防病效果[J]. 生物技术通报, 2022, 38(6): 136-146. |
| [10] | 严聪文, 苏代发, 代庆忠, 张振荣, 田云霞, 董琼娥, 周文星, 陈杉艳, 童江云, 崔晓龙. 草莓病害的生物防治研究进展[J]. 生物技术通报, 2022, 38(12): 73-87. |
| [11] | 伊帕热·帕尔哈提, 祖力胡玛尔·肉孜, 田永芝, 朱艳蕾, 李远婷, 马晓林. 荒漠植物内生菌多样性及其增强农作物抗旱和耐盐性的研究进展[J]. 生物技术通报, 2022, 38(12): 88-99. |
| [12] | 舒洁, 张仁军, 梁应冲, 陈雅琼, 张娟, 郭建, 陈穗云. 植物源与微生物源生物制剂复配防治根结线虫病[J]. 生物技术通报, 2021, 37(7): 164-174. |
| [13] | 张洁, 夏明聪, 朱文倩, 梁娟, 孙润红, 徐文, 武超, 杨丽荣. 蔬菜根结线虫生防芽胞杆菌的筛选及作用机理研究[J]. 生物技术通报, 2021, 37(7): 175-182. |
| [14] | 陈立杰, 杨帆, 范海燕, 赵迪, 王媛媛, 朱晓峰, 刘晓宇, 段玉玺. 非编码RNA在生防菌-植物线虫-寄主互作中的研究进展[J]. 生物技术通报, 2021, 37(7): 65-70. |
| [15] | 武杞蔓, 张金梅, 李玥莹, 张颖. 有益微生物菌肥对农作物的作用机制研究进展[J]. 生物技术通报, 2021, 37(5): 221-230. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||