








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 148-158.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1045
李慧1( ), 文钰芳1, 王悦1, 纪超1, 石国优1, 罗英2, 周勇1, 李志敏1, 吴晓玉1, 杨有新2(
), 文钰芳1, 王悦1, 纪超1, 石国优1, 罗英2, 周勇1, 李志敏1, 吴晓玉1, 杨有新2( ), 刘建萍1(
), 刘建萍1( )
)
收稿日期:2023-11-07
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
刘建萍,女,博士,副教授,研究方向:植物分子生物学;E-mail: JianpingLiu@jxau.edu.cn;作者简介:李慧,女,硕士研究生,研究方向:植物分子生物学;E-mail: lh15736630394@163.com文钰芳为本文共同第一作者
基金资助:
LI Hui1( ), WEN Yu-fang1, WANG Yue1, JI Chao1, SHI Guo-you1, LUO Ying2, ZHOU Yong1, LI Zhi-min1, WU Xiao-yu1, YANG You-xin2(
), WEN Yu-fang1, WANG Yue1, JI Chao1, SHI Guo-you1, LUO Ying2, ZHOU Yong1, LI Zhi-min1, WU Xiao-yu1, YANG You-xin2( ), LIU Jian-ping1(
), LIU Jian-ping1( )
)
Received:2023-11-07
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】研究辣椒光敏色素互作因子4(phytochrome-interacting factor 4, PIF4)响应盐胁迫的分子机制,为探究辣椒响应盐胁迫的分子机理和选育耐盐品种提供理论基础。【方法】以‘Zunla-1’cDNA为模板克隆CaPIF4,利用生物信息软件分析该基因所编码蛋白的理化性质,实时荧光定量PCR(RT-qPCR)技术和病毒诱导基因沉默(virus induced gene silencing, VIGS)技术研究盐胁迫处理后CaPIF4表达模式及在盐胁迫中的作用。【结果】CaPIF4在辣椒根、茎、叶、花和果实各组织均有表达,但在叶片中表达水平最高;随着300 mmol/L NaCl处理时间的增加,CaPIF4表达量升高,其中,在处理2 h后表达量达到最高;利用VIGS技术沉默CaPIF4,将对照组(CK)和基因瞬时沉默组(pTRV-CaPIF4)进行300 mmol/L NaCl处理后发现,基因沉默组植株与空载组(CK)相比,萎蔫程度更为严重,说明基因瞬时沉默组(pTRV-CaPIF4)植株耐盐性降低;亚细胞定位结果显示,CaPIF4定位于细胞核;酵母转录活性分析发现CaPIF4具有转录激活活性。高盐胁迫下,辣椒叶片过氧化氢含量升高,过氧化物酶活性升高。染色结果显示,沉默CaPIF4后,导致H2O2和O2-含量显著升高。【结论】CaPIF4作为转录因子可能参与调控辣椒的盐耐 受性。
李慧, 文钰芳, 王悦, 纪超, 石国优, 罗英, 周勇, 李志敏, 吴晓玉, 杨有新, 刘建萍. 盐胁迫下辣椒CaPIF4的表达特性与功能分析[J]. 生物技术通报, 2024, 40(4): 148-158.
LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress[J]. Biotechnology Bulletin, 2024, 40(4): 148-158.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| CaPIF4-F | ATGAATCCTTGTCTTCCTG |
| CaPIF4-R | TCAAACCTGAGAAGCACC |
| CaPIF4-qPCR-F | TGGTTGATCCATCAATCCCTT |
| CaPIF4-qPCR-R | CAAACCTGAGAAGCACCCTGA |
| CaPIF4-VIGS-F | TGAGTAAGGTTACCGAATTCATGAATCCTTGTC- TTCCTGAAT |
| CaPIF4-VIGS-R | GTGAGCTCGGTACCGGATCCACAAAATACTTTG- TCAAATGAAT |
| pGBKT7-CaPIF4-F | ATGGCCATGGAGGCCGAATTCATGAATCCTTGT- CTTCCTGAATGG |
| pGBKT7-CaPIF4-R | CCGCTGCAGGTCGACGGATCCAACCTGAGAAGCACCCTG |
| p3301-CaPIF4-F | ACGGGGGACTCTTGACCATGGATGAATCCTTG- TCTTCCTGAATGG |
| p3301-CaPIF4-F | GCCCTTGCTCACCATCCATGGAACCTGAGAAGCACCCTG |
| CaACTIN-F | AGGGATGGGTCAAAAGGATGC |
| CaACTIN-F | GAGACAACACCGCCTGAATAGC |
表1 引物列表
Table1 List of primers
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| CaPIF4-F | ATGAATCCTTGTCTTCCTG |
| CaPIF4-R | TCAAACCTGAGAAGCACC |
| CaPIF4-qPCR-F | TGGTTGATCCATCAATCCCTT |
| CaPIF4-qPCR-R | CAAACCTGAGAAGCACCCTGA |
| CaPIF4-VIGS-F | TGAGTAAGGTTACCGAATTCATGAATCCTTGTC- TTCCTGAAT |
| CaPIF4-VIGS-R | GTGAGCTCGGTACCGGATCCACAAAATACTTTG- TCAAATGAAT |
| pGBKT7-CaPIF4-F | ATGGCCATGGAGGCCGAATTCATGAATCCTTGT- CTTCCTGAATGG |
| pGBKT7-CaPIF4-R | CCGCTGCAGGTCGACGGATCCAACCTGAGAAGCACCCTG |
| p3301-CaPIF4-F | ACGGGGGACTCTTGACCATGGATGAATCCTTG- TCTTCCTGAATGG |
| p3301-CaPIF4-F | GCCCTTGCTCACCATCCATGGAACCTGAGAAGCACCCTG |
| CaACTIN-F | AGGGATGGGTCAAAAGGATGC |
| CaACTIN-F | GAGACAACACCGCCTGAATAGC |
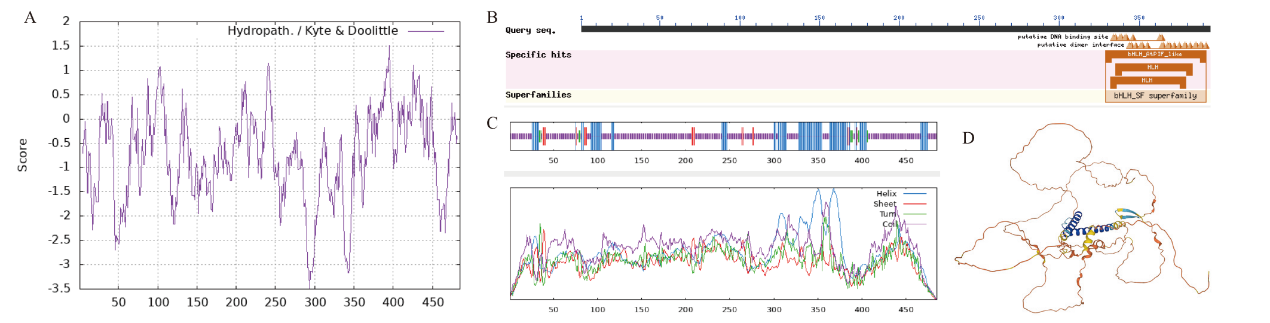
图2 CaPIF4的序列分析 A:CaPIF4的亲水性分析;B:CaPIF4保守结构域分析;C:CaPIF4二级结构;D:CaPIF4三级结构
Fig. 2 Sequence analysis of CaPIF4 A: Hydrophilicity analysis of CaPIF4 protein. B: Conserved domain analysis of CaPIF4. C: Secondary structure of CaPIF4 protein. D: Tertiary structure of CaPIF4 protein
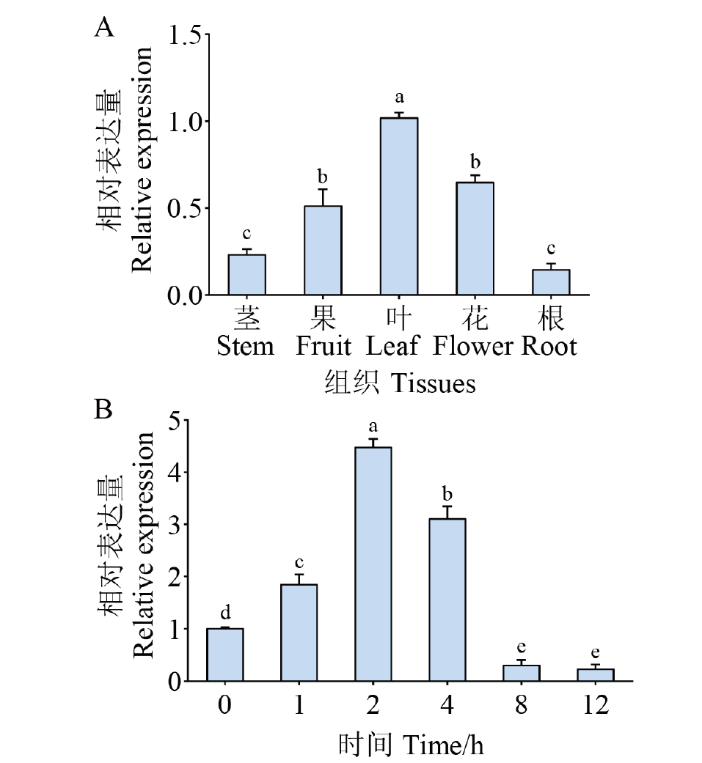
图3 CaPIF4的表达模式分析 A:不同组织中CaPIF4的表达分析;B:盐胁迫下CaPIF4在叶中的表达分析。平均值和标准差分别由3个独立实验得到,每个实验3株植物。不同小写字母表示差异有统计学意义,P≤0.05
Fig. 3 Expression pattern analysis of CaPIF4 A: Expression analysis of CaPIF4 in different tissues. B: Expression analysis of CaPIF4 in the leaves under salt stress. Mean and SD values were obtained from three independent experiments, with 3 plants per experiments. The different letters indicate significant differences at P ≤0.05

图4 pTRV2-CaPIF4载体的构建 A:pTRV2-CaPIF4载体的酶切验证;B:pTRV2-CaPIF4载体序列比对;M:BM200 DNA marker;1-3:3个重复
Fig. 4 Construction of pTRV2-CaPIF4 vector A: Enzyme digesting verification of pTRV2-CaPIF4 vector; B: pTRV2-CaPIF4 vector sequence alignment; M: BM2000 DNA marker; 1-3: three repeats
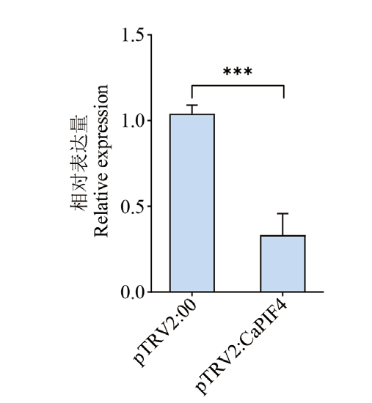
图6 CaPIF4沉默效率检测 图中误差线表示标准偏差,星号代表显著差异(*P<0.05,** P<0.01,***P<0.001),下同
Fig. 6 Detection of CaPIF4 gene silencing efficiency The error line in the figure refers to the standard deviation. * indicate statistical significance (*P<0.05,**P<0.01,***P<0.001 ) compared to control. The same below
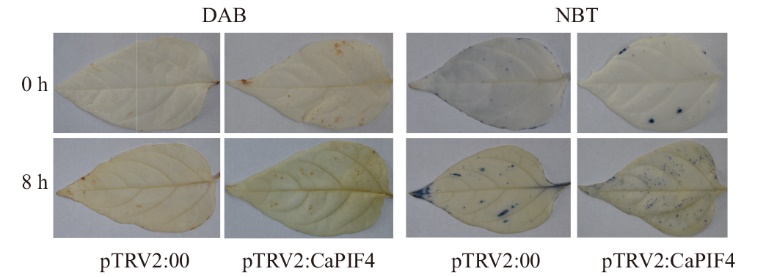
图10 盐胁迫前后对照组和VIGS沉默植株第3、4叶片中ROS积累情况 A:3,3' -二氨基联苯胺(DAB)染色;B:氮蓝四唑(NBT)染色
Fig. 10 ROS accumulation in the control group and VIGS silenced plants in the 3rd and 4th leaves before and after salt stress A: 3,3'-Diaminobenzidine (DAB) staining. B: Nitro-blue tetrazolium (NBT) staining
| [1] |
Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub[J]. Trends Plant Sci, 2011, 16(1): 19-28.
doi: 10.1016/j.tplants.2010.08.003 pmid: 20833098 |
| [2] |
Lee N, Choi G. Phytochrome-interacting factor from Arabidopsis to liverwort[J]. Curr Opin Plant Biol, 2017, 35: 54-60.
doi: 10.1016/j.pbi.2016.11.004 URL |
| [3] |
Pham VN, Kathare PK, Huq E. Phytochromes and phytochrome interacting factors[J]. Plant Physiol, 2018, 176(2): 1025-1038.
doi: 10.1104/pp.17.01384 pmid: 29138351 |
| [4] |
Inoue K, Nishihama R, Kataoka H, et al. Phytochrome signaling is mediated by phytochrome interacting factor in the liverwort Marchantia polymorpha[J]. Plant Cell, 2016, 28(6): 1406-1421.
doi: 10.1105/tpc.15.01063 URL |
| [5] |
Possart A, Xu TF, Paik I, et al. Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants[J]. Plant Cell, 2017, 29(2): 310-330.
doi: 10.1105/tpc.16.00388 URL |
| [6] |
Nakamura Y, Kato T, Yamashino T, et al. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa[J]. Biosci Biotechnol Biochem, 2007, 71(5): 1183-1191.
doi: 10.1271/bbb.60643 URL |
| [7] |
Shi QB, Zhang HS, Song XY, et al. Functional characterization of the maize phytochrome-interacting factors PIF4 and PIF5[J]. Front Plant Sci, 2018, 8: 2273.
doi: 10.3389/fpls.2017.02273 URL |
| [8] |
Wu GX, Zhao YP, Shen RX, et al. Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis[J]. Plant Physiol, 2019, 181(2): 789-803.
doi: 10.1104/pp.19.00239 pmid: 31350363 |
| [9] | Zheng PF, Wang X, Yang YY, et al. Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica[J]. Int J Mol Sci, 2020, 21(19): 7350. |
| [10] | Rosado D, Gramegna G, Cruz A, et al. Phytochrome interacting factors(PIFs)in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes[J]. PLoS One, 2016, 11(11): e0165929. |
| [11] |
Ding JH, Zhang B, Li Y, et al. Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 modulate seasonal growth in trees[J]. New Phytol, 2021, 232(6): 2339-2352.
doi: 10.1111/nph.17350 pmid: 33735450 |
| [12] |
Li WQ, Liu Y, Wang WP, et al. Phytochrome-interacting factor(PIF)in rapeseed(Brassica napus L.): Genome-wide identification, evolution and expression analyses during abiotic stress, light quality and vernalization[J]. Int J Biol Macromol, 2021, 180: 14-27.
doi: 10.1016/j.ijbiomac.2021.03.055 URL |
| [13] |
Leivar P, Monte E. PIFs: systems integrators in plant development[J]. Plant Cell, 2014, 26(1): 56-78.
doi: 10.1105/tpc.113.120857 URL |
| [14] |
Sakuraba Y, Bülbül S, Piao WL, et al. Arabidopsis early flowering3 increases salt tolerance by suppressing salt stress response pathways[J]. Plant J, 2017, 92(6): 1106-1120.
doi: 10.1111/tpj.2017.92.issue-6 URL |
| [15] |
Gao Y, Wu MQ, Zhang MJ, et al. Roles of a maize phytochrome-interacting factors protein ZmPIF3 in regulation of drought stress responses by controlling stomatal closure in transgenic rice without yield penalty[J]. Plant Mol Biol, 2018, 97(4/5): 311-323.
doi: 10.1007/s11103-018-0739-4 |
| [16] |
Ma L, Han R, Yang YQ, et al. Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance[J]. Plant Cell, 2023, 35(8): 2997-3020.
doi: 10.1093/plcell/koad117 URL |
| [17] |
Mo WP, Tang WJ, Du YX, et al. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress[J]. Plant Physiol, 2020, 184(1): 506-517.
doi: 10.1104/pp.20.00024 pmid: 32581115 |
| [18] |
Yang YX, Guang YL, Wang F, et al. Characterization of phytochrome-interacting factor genes in pepper and functional analysis of CaPIF8 in cold and salt stress[J]. Front Plant Sci, 2021, 12: 746517.
doi: 10.3389/fpls.2021.746517 URL |
| [19] |
Paik I, Kathare PK, Kim JI, et al. Expanding roles of PIFs in signal integration from multiple processes[J]. Mol Plant, 2017, 10(8): 1035-1046.
doi: S1674-2052(17)30198-3 pmid: 28711729 |
| [20] |
Bu QY, Zhu L, Huq E. Multiple kinases promote light-induced degradation of PIF1[J]. Plant Signal Behav, 2011, 6(8): 1119-1121.
doi: 10.4161/psb.6.8.16049 pmid: 21758014 |
| [21] |
Al-Sady B, Ni WM, Kircher S, et al. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation[J]. Mol Cell, 2006, 23(3): 439-446.
doi: 10.1016/j.molcel.2006.06.011 pmid: 16885032 |
| [22] |
Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis[J]. Plant J, 2005, 44(6): 1023-1035.
doi: 10.1111/j.1365-313X.2005.02606.x pmid: 16359394 |
| [23] |
Ni WM, Xu SL, Chalkley RJ, et al. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis[J]. Plant Cell, 2013, 25(7): 2679-2698.
doi: 10.1105/tpc.113.112342 URL |
| [24] | Paik I, Huq E. Rapid examination of phytochrome-phytochrome interacting factor(PIF)interaction by in vitro coimmunoprecipitation assay[J]. Methods Mol Biol, 2019, 2026: 21-28. |
| [25] |
Ni WM, Xu SL, González-Grandío E, et al. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3[J]. Nat Commun, 2017, 8: 15236.
doi: 10.1038/ncomms15236 pmid: 28492231 |
| [26] |
Bernardo-García S, de Lucas M, Martínez C, et al. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth[J]. Genes Dev, 2014, 28(15): 1681-1694.
doi: 10.1101/gad.243675.114 URL |
| [27] |
Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis[J]. Plant Physiol, 2011, 156(1): 357-372.
doi: 10.1104/pp.111.172684 URL |
| [28] |
Lorrain S, Allen T, Duek PD, et al. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors[J]. Plant J, 2008, 53(2): 312-323.
doi: 10.1111/j.1365-313X.2007.03341.x pmid: 18047474 |
| [29] |
Huang GT, Ma SL, Bai LP, et al. Signal transduction during cold, salt, and drought stresses in plants[J]. Mol Biol Rep, 2012, 39(2): 969-987.
doi: 10.1007/s11033-011-0823-1 URL |
| [30] | Ma YL, Cao J, He JH, et al. Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses[J]. Int J Mol Sci, 2018, 19(11): 3643. |
| [31] |
Zhu JK. Salt and drought stress signal transduction in plants[J]. Annu Rev Plant Biol, 2002, 53: 247-273.
doi: 10.1146/arplant.2002.53.issue-1 URL |
| [32] | 李俊贞, 何乐祖, 赵春梅, 等. 盐胁迫对黄果厚壳桂幼苗荧光和生理特性的影响[J]. 山西农业科学, 2021, 49(8): 919-923. |
| Li JZ, He LZ, Zhao CM, et al. Effects of salt stress on the photosynthetic and physiological characteristics of cryptocarya concinna seedlings[J]. J Shanxi Agric Sci, 2021, 49(8): 919-923. | |
| [33] | 付丽, 刘加珍, 陶宝先, 等. 盐生植物对盐渍土壤环境的适应机制研究综述[J]. 江苏农业科学, 2021, 49(15): 32-39. |
| Fu L, Liu JZ, Tao BX, et al. A review on the adaptationmechanism of halophytes to saline soil environment[J]. J Jiangsu Agric Sci, 2021, 49(15): 32-39. | |
| [34] |
Huang RD. Research progress on plant tolerance to soil salinity and alkalinity in sorghum[J]. J Integr Agric, 2018, 17(4): 739-746.
doi: 10.1016/S2095-3119(17)61728-3 URL |
| [35] | 秦峰梅, 张红香, 武祎, 等. 盐胁迫对黄花苜蓿发芽及幼苗生长的影响[J]. 草业学报, 2010, 19(4): 71-78. |
| Qin FM, Zhang HX, Wu Y, et al. Effects of salt stress on germination and seedling growth of Medicago falcata[J]. Acta Prataculturae Sin, 2010, 19(4): 71-78. | |
| [36] |
时振振, 李胜, 杨柯, 等. 盐胁迫下豌豆幼苗对内外源NO的生理生化响应[J]. 草业学报, 2014, 23(5): 193-200.
doi: 10.11686/cyxb20140522 |
| Shi ZZ, Li S, Yang K, et al. Physiological and biochemical response of pea seedlings to endogenous and exogenous NO under salt stress[J]. Acta Prataculturae Sin, 2014, 23(5): 193-200. | |
| [37] | 孙璐, 黄瑞冬. 高粱幼苗保护酶系统对盐胁迫的初期响应[J]. 沈阳农业大学学报, 2014, 45(2): 134-137. |
| Sun L, Huang RD. Responses to salt stress of protective enzyme system in sorghum seedlings[J]. J Shenyang Agric Univ, 2014, 45(2): 134-137. | |
| [38] | 田晓艳, 刘延吉, 郭迎春. 盐胁迫对NHC牧草Na+、K+、Pro、可溶性糖及可溶性蛋白的影响[J]. 草业科学, 2008, 25(10): 34-38. |
| Tian XY, Liu YJ, Guo YC. Effect of salt stress on Na+, K+, Pro, soluble sugar and soluble protein of NHC[J]. Pratacultural Sci, 2008, 25(10): 34-38. | |
| [39] | Feng XJ, Li JR, Qi SL, et al. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis[J]. Proc Natl Acad Sci USA. 2016, 113(51): E8335-E8343. |
| [40] | Seong ES, Cho HS, Choi D, et al. Tomato plants overexpressing CaKR1 enhanced tolerance to salt and oxidative stress[J]. Biochem Biophys Res Commun, 2007 ; 363(4): 983-988. |
| [41] |
宋阳, 崔晓晗, 张明, 等. 盐胁迫对番茄幼苗生理特性及离子分布的影响[J]. 北方农业学报, 2019, 47(4): 115-121.
doi: 10.3969/j.issn.2096-1197.2019.04.20 |
| Song Y, Cui XH, Zhang M, et al. Effects of salt stress on physiological characteristics and ion distribution of tomato seedlings[J]. J North Agric, 2019, 47(4): 115-121. |
| [1] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [2] | 郭纯, 宋桂梅, 闫艳, 邸鹏, 王英平. 西洋参bZIP基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2024, 40(4): 167-178. |
| [3] | 高玉坤, 张建东, 杨溥原, 陈东明, 王志博, 田颐瑾, 崔江慧, 常金华. 高粱根际土壤细菌群落对盐胁迫的响应[J]. 生物技术通报, 2024, 40(4): 203-216. |
| [4] | 高志伟, 魏明, 于祖隆, 伍国强, 魏俊龙. 耐盐植物促生菌W-1鉴定及其对红豆草耐盐性的影响[J]. 生物技术通报, 2024, 40(4): 217-227. |
| [5] | 毛立杰, 梁晓, 刘迎, 伍春玲, 韩晓燕, 陈青. CMV通过影响效应因子MpC002的表达干预桃蚜种群增长的机制[J]. 生物技术通报, 2024, 40(4): 271-277. |
| [6] | 吴星星, 洪海波, 甘志承, 李瑞宁, 黄先忠. 辣椒CaPI的克隆与功能分析[J]. 生物技术通报, 2024, 40(3): 193-201. |
| [7] | 谢倩, 江来, 贺进, 刘玲玲, 丁明月, 陈清西. 不同鲜食品质橄榄果实转录组测序及酚类代谢途径相关调控基因挖掘[J]. 生物技术通报, 2024, 40(3): 215-228. |
| [8] | 沈天虹, 齐孝博, 赵瑞丰, 马欣荣. 微藻盐胁迫响应分子机制研究进展[J]. 生物技术通报, 2024, 40(3): 89-99. |
| [9] | 陈艳梅. 蛋白质翻译后修饰之间的互作关系及其协同调控机理[J]. 生物技术通报, 2024, 40(2): 1-8. |
| [10] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [11] | 路喻丹, 刘晓驰, 冯新, 陈桂信, 陈义挺. 猕猴桃BBX基因家族成员鉴定与转录特征分析[J]. 生物技术通报, 2024, 40(2): 172-182. |
| [12] | 李昊, 伍国强, 魏明, 韩悦欣. 甜菜BvBADH基因家族全基因组鉴定及其高盐胁迫下的表达分析[J]. 生物技术通报, 2024, 40(2): 233-244. |
| [13] | 徐扬, 张瑞英, 戴良香, 张冠初, 丁红, 张智猛. 盐胁迫下氮素对花生种子萌发和种子际细菌菌群结构的调控[J]. 生物技术通报, 2024, 40(2): 253-265. |
| [14] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [15] | 王雨晴, 马子奇, 侯嘉欣, 宗钰琪, 郝晗睿, 刘国元, 魏辉, 连博琳, 陈艳红, 张健. 盐胁迫下植物根系分泌物的成分分析与生态功能研究进展[J]. 生物技术通报, 2024, 40(1): 12-23. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||