








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 90-98.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0908
邹修为1( ), 岳佳妮1, 李志宇2, 戴良英1, 李魏1(
), 岳佳妮1, 李志宇2, 戴良英1, 李魏1( )
)
收稿日期:2023-09-21
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
李魏,男,博士,教授,研究方向:植物与微生物互作;E-mall: liwei350551@163.com作者简介:邹修为,男,硕士研究生,研究方向:植物与微生物互作;E-mall: zouxiuwei0520@163.com
基金资助:
ZOU Xiu-wei1( ), YUE Jia-ni1, LI Zhi-yu2, DAI Liang-ying1, LI Wei1(
), YUE Jia-ni1, LI Zhi-yu2, DAI Liang-ying1, LI Wei1( )
)
Received:2023-09-21
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】水稻在生长发育过程中,常会受到各种非生物胁迫而严重影响产量。热激转录因子作为植物抗逆过程中的一个重要元件,通过调控一系列胁迫响应基因的表达以提高植物抗逆性。探究水稻热激转录因子OsHsfA2b调控非生物胁迫的功能与初步机理,为培育水稻抗逆新品种提供了优异基因资源和理论支撑。【方法】通过构建OsHsfA2b过量表达和RNAi的转基因水稻,分别观察水稻幼苗在高温、低温、干旱和高盐处理后的抗逆表型,统计存活率。同时,在逆境胁迫处理后,通过DAB染色检测水稻叶片活性氧(ROS)含量及RT-qPCR检测抗氧化途径相关基因OsSOD和OsCAT的表达量,分析OsHsfA2b对抗氧化途径的调控作用。【结果】在高温、低温、干旱和高盐等非生物胁迫条件下,水稻热激转录因子OsHsfA2b被显著诱导表达。与野生型NPB相比,OsHsfA2b过量表达转基因水稻对非生物胁迫的抗性明显增强,植株损伤程度较轻,存活率提高。相反,OsHsfA2b-RNAi水稻对非生物胁迫更加敏感,植株损伤严重,存活率降低。此外,在非生物胁迫条件下,OsHsfA2b过量表达转基因水稻比NPB和OsHsfA2b-RNAi水稻体内活性氧的含量减少,并且OsHsfA2b显著诱导了抗氧化途径相关基因OsSOD和OsCAT的表达,OsHsfA2b参与抑制水稻体内活性氧的积累以降低逆境胁迫诱导的活性氧大量积累对植株的伤害。【结论】非生物胁迫下,水稻热激转录因子OsHsfA2b被显著诱导表达,可通过抗氧化途径正调控水稻对逆境胁迫的抗性。
邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98.
ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses[J]. Biotechnology Bulletin, 2024, 40(2): 90-98.
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| UBQ-F | AACCAGCTGAGGCCCAAGA |
| UBQ-R | ACGATTGATTTAACCAGTCCATGA |
| OsHsfA2b-F | GCCTTGTTTGATCCCCAGGA |
| OsHsfA2b-R | TTGTCGTTCAACTCCCCCTG |
| OsSOD-F | AACAAAGGCAGGGCTGTAGA |
| OsSOD-R | TAAGGTTCCAGGGCATCAAG |
| OsCAT-F | TGAAGCCAAGCATGTGAAGA |
| OsCAT-R | GCCCAACGACAACAGAAGAT |
表1 实时荧光定量PCR引物
Table 1 Primers for RT-qPCR
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| UBQ-F | AACCAGCTGAGGCCCAAGA |
| UBQ-R | ACGATTGATTTAACCAGTCCATGA |
| OsHsfA2b-F | GCCTTGTTTGATCCCCAGGA |
| OsHsfA2b-R | TTGTCGTTCAACTCCCCCTG |
| OsSOD-F | AACAAAGGCAGGGCTGTAGA |
| OsSOD-R | TAAGGTTCCAGGGCATCAAG |
| OsCAT-F | TGAAGCCAAGCATGTGAAGA |
| OsCAT-R | GCCCAACGACAACAGAAGAT |
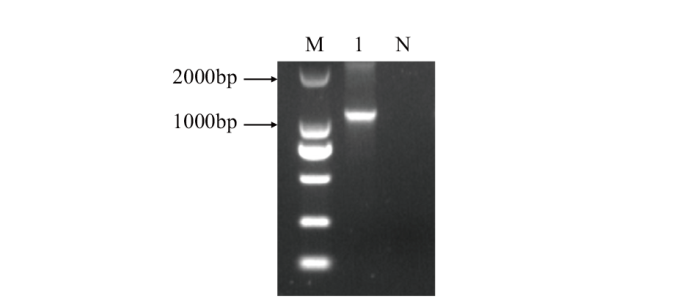
图1 OsHsfA2b的CDS扩增电泳图 M:Marker;N:阴性对照;1:OsHsfA2b CDS全长扩增片段
Fig. 1 Electropherogram of OsHsfA2b gene CDS amplification M: Marker; N: negative control; 1: OsHsfA2b full-length CDS amplified fragment

图2 OsHsfA2b的蛋白结构 结构图上方数字表示氨基酸位置。DBD:DNA结合结构域;OD:寡聚化结构域;NLS:核定位信号;CTAD:C端转录激活结构域;NES:C端核输出信号
Fig. 2 Protein structure of OsHsfA2b The number above the structural diagram represents the position of amino acids. DBD: DNA binding domain; OD: oligomerization domain; NLS: nuclear localization signal; CTAD: C-terminal transcriptional activation domain; NES: nuclear export signal

图3 OsHsfA2b-OX和OsHsfA2b-RNAi转基因植株的鉴定 A:OsHsfA2b-OX转基因植株中OsHsfA2b的表达情况;B:OsHsfA2b-RNAi植株中OsHsfA2b的表达情况
Fig. 3 Identification of OsHsfA2b-OX and OsHsfA2b-RNAi transgenic plants A: OsHsfA2b gene expression of OsHsfA2b-OX transgenic plants. B: OsHsfA2b gene expression of OsHsfA2b-RNAi transgenic plants
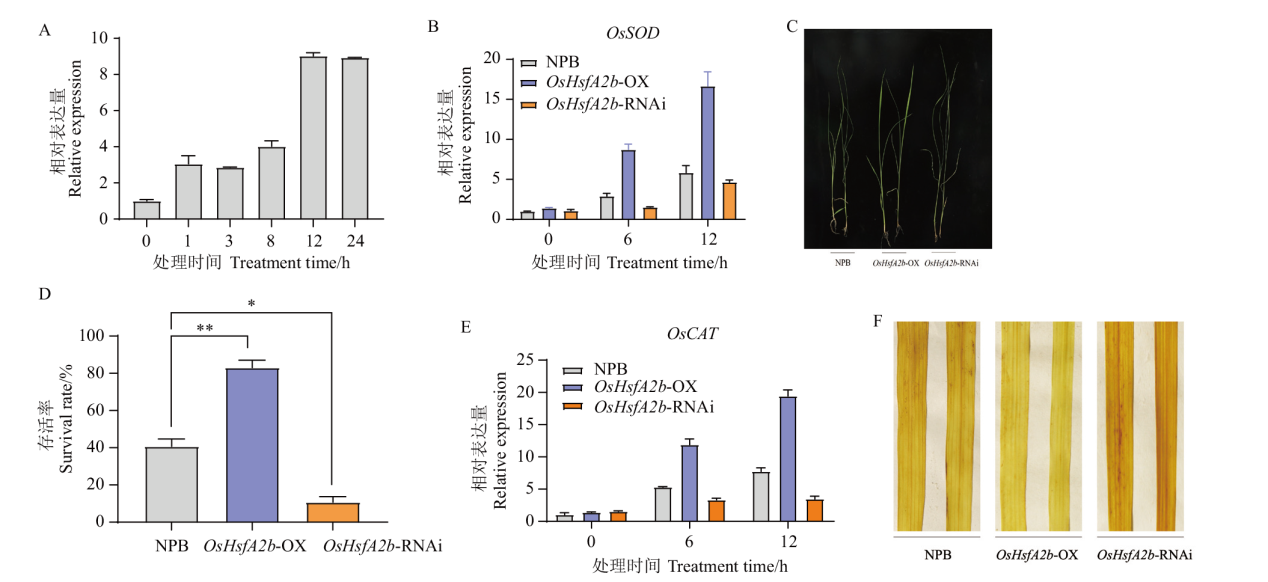
图4 高温胁迫下转基因植株的表型和OsHsfA2b的表达 A :高温胁迫(42℃)NPB 植株中OsHsfA2b 的表达量;B :高温处理后OsSOD 的表达;C :高温处理2 d 后植株的表型; D : 存活率统计;E:高温处理后OsCAT 的表达;F :活性氧含量的检测。* 表示差异显著(P<0.05),** 表示差异极显著(P<0.01),下同
Fig. 4 Phenotypes and OsHsfA2b expressions of transgenic plants under high temperature stress A: Expression of OsHsfA2b in NPB under high temperature stress(42℃). B: Expression of OsSOD after high temperature treatment. C: Phenotypes of plants after high temperature treatment for 2 d after high temperature treatment. D: Survival rate statistic. E: Expression of OsCAT after high temperature treatment. F: Determination of reactive oxygen species content. * indicates significant difference(P<0.05), and **indicates extremely significant difference(P<0.01). The same below

图5 低温胁迫下转基因植株的表型和OsHsfA2b的表达 A:低温胁迫(4℃)OsHsfA2b的表达量;B:低温处理2 d后的表型;C:存活率统计;D-E:低温处理后OsSOD和OsCAT的表达量
Fig. 5 Phenotypes and OsHsfA2b expressions of transgenic plants under low temperature stress A: Expression of OsHsfA2b under low temperature stress(4℃). B: Phenotypes of plants after low temperature treatment for 2 d. C: Survival rate statistic. D-E: Expression of OsSOD and OsCAT after low temperature treatment

图6 干旱胁迫下转基因植株的表型和OsHsfA2b的表达 A:干旱处理OsHsfA2b的表达量;B:干旱处理1 d后的表型;C:存活率统计;D-E:干旱处理后OsSOD和OsCAT的表达量
Fig. 6 Phenotypes and OsHsfA2b expressions of transgenic plants under drought stress A: Expression of OsHsfA2b under drought treatment. B: Phenotypes of plants after drought treatment for 1 d. C: Survival rate statistic. D-E: Expression of OsSOD and OsCAT after drought treatment
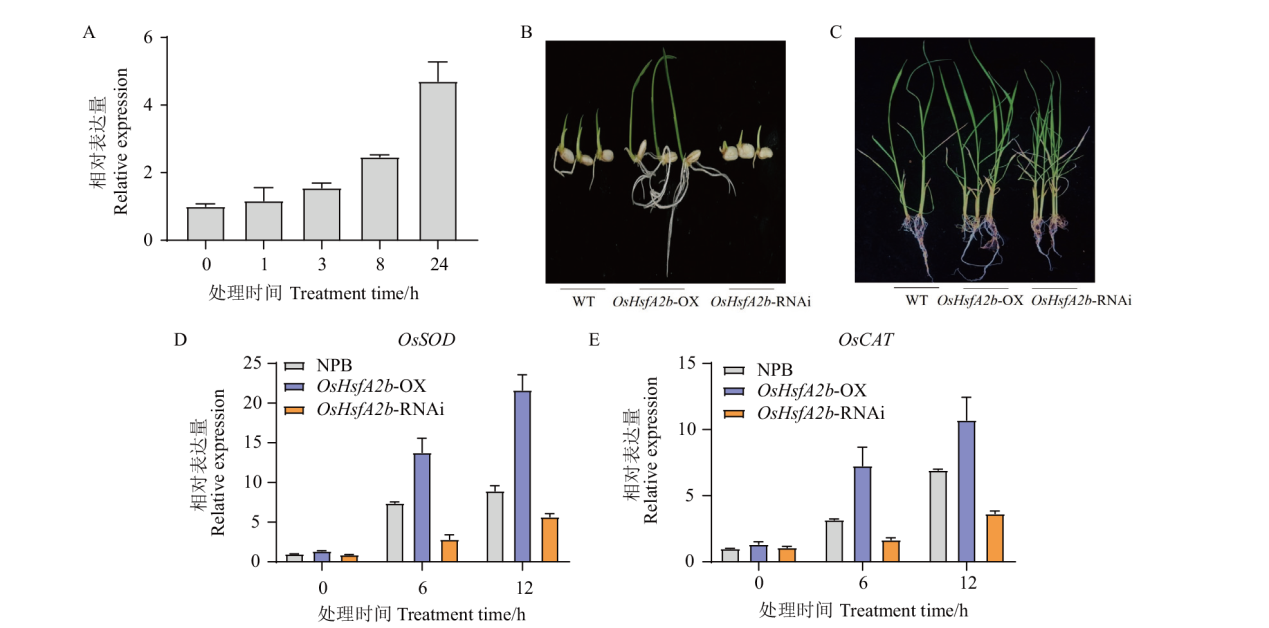
图7 高盐胁迫下转基因植株的表型和OsHsfA2b的表达 A:150 mmol/L NaCl处理后OsHsfA2b的表达量;B:150 mmol/L NaCl处理后种子萌发5 d的表型;C:150 mmol/L NaCl处理2 d后的表型;D-E:高盐处理后OsSOD和OsCAT的表达量
Fig. 7 Phenotypes and OsHsfA2b expressions of transgenic plants under high salt stress A: Expression of OsHsfA2b under 150 mmol/L NaCl. B: Phenotypes of seedlings geminated after 150 mmol/L NaCl for 5 d. C: Phenotypes of plants under 150 mmol/L NaCl for 2 d. D-E: Expression of OsSOD and OsCAT after high salt treatment
| [1] |
Haider S, Raza A, Iqbal J, et al. Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal[J]. Mol Biol Rep, 2022, 49(6): 5771-5785.
doi: 10.1007/s11033-022-07190-x |
| [2] |
Ohama N, Sato H, Shinozaki K, et al. Transcriptional regulatory network of plant heat stress response[J]. Trends Plant Sci, 2017, 22(1): 53-65.
doi: S1360-1385(16)30126-1 pmid: 27666516 |
| [3] |
Li PS, Yu TF, He GH, et al. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses[J]. BMC Genomics, 2014, 15(1): 1009.
doi: 10.1186/1471-2164-15-1009 |
| [4] |
Nover L, Bharti K, Döring P, et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?[J]. Cell Stress Chaperones, 2001, 6(3): 177-189.
doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2 URL |
| [5] |
Baniwal SK, Bharti K, Chan KY, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors[J]. J Biosci, 2004, 29(4): 471-487.
doi: 10.1007/BF02712120 URL |
| [6] |
Rehman A, Atif RM, Azhar MT, et al. Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons(Gossypium spp.)[J]. Int J Biol Macromol, 2021, 182: 1507-1527.
doi: 10.1016/j.ijbiomac.2021.05.016 pmid: 33965497 |
| [7] |
Lin YX, Jiang HY, Chu ZX, et al. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize[J]. BMC Genomics, 2011, 12: 76.
doi: 10.1186/1471-2164-12-76 |
| [8] | 张楠, 王映红, 王志敏, 等. 植物热激转录因子家族的研究进展[J]. 生物工程学报, 2021, 37(4): 1155-1167. |
| Zhang N, Wang YH, Wang ZM, et al. Heat shock transcription factor family in plants: a review[J]. Chin J Biotechnol, 2021, 37(4): 1155-1167. | |
| [9] | 焦淑珍, 姚文孔, 张宁波, 等. 园艺植物热激转录因子研究进展[J]. 果树学报, 2020, 37(3): 419-430. |
| Jiao SZ, Yao WK, Zhang NB, et al. Research progress of heat stress transcription factors(Hsfs)in horticultural plants[J]. J Fruit Sci, 2020, 37(3): 419-430. | |
| [10] |
Fragkostefanakis S, Mesihovic A, Simm S, et al. HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues[J]. Plant Physiol, 2016, 170(4): 2461-2477.
doi: 10.1104/pp.15.01913 pmid: 26917685 |
| [11] | Zhuang L, Cao W, Wang J, et al. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis[J]. Int J Mol Sci, 2018, 19(9): E2702. |
| [12] | Guo M, Liu JH, Ma X, et al. The plant heat stress transcription factors(HSFs): Structure, regulation, and function in response to abiotic stresses[J]. Front Plant Sci, 2016, 7: 114. |
| [13] |
Chan-Schaminet KY, Baniwal SK, Bublak D, et al. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression[J]. J Biol Chem, 2009, 284(31): 20848-20857.
doi: 10.1074/jbc.M109.007336 pmid: 19491106 |
| [14] |
Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development[J]. Plant Physiol, 2013, 163(1): 276-290.
doi: 10.1104/pp.113.221168 URL |
| [15] |
Poonia AK, Mishra SK, Sirohi P, et al. Overexpression of wheat transcription factor(TaHsfA6b)provides thermotolerance in barley[J]. Planta, 2020, 252(4): 53.
doi: 10.1007/s00425-020-03457-4 pmid: 32945950 |
| [16] |
Prieto-Dapena P, Castaño R, Almoguera C, et al. Improved resistance to controlled deterioration in transgenic seeds[J]. Plant Physiol, 2006, 142(3): 1102-1112.
doi: 10.1104/pp.106.087817 pmid: 16998084 |
| [17] |
Liu H, Zhang Y, Lu S, et al. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis[J]. New Phytol, 2021, 231(2): 646-660.
doi: 10.1111/nph.v231.2 URL |
| [18] |
Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants?[J]. Ann Bot, 2006, 98(2): 279-288.
doi: 10.1093/aob/mcl107 URL |
| [19] |
Pérez-Salamó I, Papdi C, Rigó G, et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6[J]. Plant Physiol, 2014, 165(1): 319-334.
doi: 10.1104/pp.114.237891 pmid: 24676858 |
| [20] |
Hwang SM, Kim DW, Woo MS, et al. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions[J]. Plant Cell Environ, 2014, 37(5): 1202-1222.
doi: 10.1111/pce.2014.37.issue-5 URL |
| [21] |
Shah Z, Iqbal A, Khan FU, et al. Genetic manipulation of pea(Pisum sativum L.)with Arabidopsis's heat shock factor HsfA1d improves ROS scavenging system to confront thermal stress[J]. Genet Resour Crop Evol, 2020, 67(8): 2119-2127.
doi: 10.1007/s10722-020-00966-9 |
| [22] |
Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth[J]. J Exp Bot, 2007, 58(12): 3373-3383.
doi: 10.1093/jxb/erm184 URL |
| [23] |
Mittal D, Madhyastha DA, Grover A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice: combination of stresses evokes different transcriptional changes as against stresses applied individually[J]. Plant Sci, 2012, 197: 102-113.
doi: 10.1016/j.plantsci.2012.09.008 pmid: 23116677 |
| [24] | 刘爱玲. 水稻 OsHsfAs 和 OsCH2基因的功能研究[D]. 长沙: 湖南农业大学, 2010. |
| Liu AL. Study on the functions of OsHsfAs and OsCH2 genes in rice[D]. Changsha: Hunan Agricultural University, 2010. | |
| [25] |
Chauhan H, Khurana N, Agarwal P, et al. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment[J]. PLoS One, 2013, 8(11): e79577.
doi: 10.1371/journal.pone.0079577 URL |
| [26] |
Mittler R. Oxidative stress, antioxidants and stress tolerance[J]. Trends Plant Sci, 2002, 7(9): 405-410.
doi: 10.1016/s1360-1385(02)02312-9 pmid: 12234732 |
| [27] |
Nishizawa A, Yabuta Y, Yoshida E, et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress[J]. Plant J, 2006, 48(4): 535-547.
doi: 10.1111/j.1365-313X.2006.02889.x pmid: 17059409 |
| [1] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [2] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [3] | 辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211. |
| [4] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| [5] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [6] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [7] | 林鑫焱, 张传忠, 戴兵, 王馨珩, 刘剑锋, 温丽, 徐兴健, 方军. 水稻穗发芽遗传与分子机制的研究进展[J]. 生物技术通报, 2024, 40(1): 24-31. |
| [8] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [9] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [10] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [11] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [12] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [13] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [14] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [15] | 周振超, 郑吉, 帅馨怡, 林泽俊, 陈红. 高通量分析人类粪便、皮肤和水环境中共享抗生素抗性基因的分布[J]. 生物技术通报, 2023, 39(7): 288-297. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||