








生物技术通报 ›› 2024, Vol. 40 ›› Issue (3): 215-228.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0747
收稿日期:2023-08-03
出版日期:2024-03-26
发布日期:2024-04-08
通讯作者:
陈清西,男,教授,研究方向:园艺植物栽培生理;E-mail: cqx0246@fafu.edu.cn作者简介:谢倩,女,博士研究生,研究方向:果树生理生化与生态;E-mail: xieq0416@163.com
基金资助:
XIE Qian( ), JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi(
), JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi( )
)
Received:2023-08-03
Published:2024-03-26
Online:2024-04-08
摘要:
【目的】 果实酚类物质类别、含量是与橄榄营养及风味密切相关的重要品质性状,探究橄榄酚类物质生物合成的分子调控机制。【方法】 以总酚含量差异显著的橄榄(低酚/高酚)为试材,分别取花后80-160 d的果实进行转录组分析,对莽草酸-水解单宁/苯丙烷-类黄酮生物合成通路差异基因进行表征,并对差异表达基因进行WGCNA分析,从中挖掘与酚类代谢途径相关的转录因子。【结果】 转录组测序共获得296 314条Unigene,其中73%的Unigene被注释到数据库;4个品种(系)橄榄成熟果共鉴定到1 628个差异表达基因(DEGs),KEGG分析显示,DEGs在酚类代谢途径的“类黄酮生物合成”通路上显著富集;进一步对果实成熟过程莽草酸-水解单宁/苯丙烷-类黄酮生物合成途径的DEGs进行表征,结合WGCNA分析中每个模块与性状的R2和P值,筛选到4个关键模块,根据模块内基因的调控关系利用MCC拓扑分析法以度值≥1挖掘模块内关键转录因子,挖掘到137个转录因子Unigene与30个酚类合成结构基因Unigene共表达,转录因子Unigene功能注释来自35个基因家族,最多的为锌指蛋白(C2C2、C3H、C2H2、PHD),其次为B3、HB、MYB、NAC基因家族。【结论】 从转录组角度初步解析了橄榄鲜食品质在酚类代谢途径的差异,同时挖掘了调控酚类代谢的差异基因,对进一步探究橄榄鲜食品质差异的分子机制提供了重要依据。
谢倩, 江来, 贺进, 刘玲玲, 丁明月, 陈清西. 不同鲜食品质橄榄果实转录组测序及酚类代谢途径相关调控基因挖掘[J]. 生物技术通报, 2024, 40(3): 215-228.
XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality[J]. Biotechnology Bulletin, 2024, 40(3): 215-228.
| 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增品种(系)Amplification variety(lines) |
|---|---|---|
| Cluster-0.119989-F | TGACTTCTGTGTCCTGCTCTT | DQc、TQc |
| Cluster-0.119989-R | GGTAGTGCCAACTGCTTCTTC | |
| Cluster-0.121834-F | CTTGACATGAATACAGCAGAGGAT | DQc、TQc |
| Cluster-0.121834-R | GCACCATCAATTTCAGCAACTT | |
| Cluster-0.121871-F | TCTGTGTCCTGCTCTTTGAAG | DQc、TQc |
| Cluster-0.121871-R | GCAACCTCAGAGATGGGAAG | |
| Cluster-0.121919-F | TAACCATAACGCCTCCATCTCT | DQc、TQc |
| Cluster-0.121919-R | TCATCTCACGAGACACAATAGACT | |
| Cluster-0.121949-F | TAACCATAACGCCTCCTTCTCT | DQc、TQc |
| Cluster-0.121949-R | GCTCCTACAATCACCACATCAG | |
| Cluster-0.155662-F | AGACTATGACGAGCAAGACAACT | DQc、TQc |
| Cluster-0.155662-R | GTTACCACACCACCGACTCT | |
| Cluster-0.119990-F | CTGTTGCCGTAACTTCACTGT | TQc |
| Cluster-0.119990-R | CTGAGAGATGGGAAGCGTTTC | |
| Cluster-0.120915-F | ATCATTCTCCAGGCATTTCTCAG | TQc |
| Cluster-0.120915-R | GGAACAGCCACAGCTTGTAT | |
| Cluster-0.121783-F | CAGGAAGTGTCAGCAACATCAA | TQc |
| Cluster-0.121783-R | CTGTTCTTCGCAAGGTTCATCT | |
| Cluster-0.121896-F | AGGAGTGAAGTGGTTGAAGAGTA | DQc |
| Cluster-0.121896-R | ACAATCATCCCAGGCACAATC | |
| Cluster-0.122437-F | TAACCATAACGCCTCCATCTCT | DQc |
| Cluster-0.122437-R | ATCAGTGTCCGCATGTGTAATC | |
| Cluster-0.122522-F | CCTCCATCTCTGCTTCTTCCT | DQc |
| Cluster-0.122522-R | TGTACCTGCGTGTCATCTCA | |
| 18S rRNA-F(内参基因) | CCTGAGAAACGGCTACCACA | DQc、TQc |
| 18S rRNA-R(内参基因) | CACCAGACTT GCCCTCCA |
表1 RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR
| 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增品种(系)Amplification variety(lines) |
|---|---|---|
| Cluster-0.119989-F | TGACTTCTGTGTCCTGCTCTT | DQc、TQc |
| Cluster-0.119989-R | GGTAGTGCCAACTGCTTCTTC | |
| Cluster-0.121834-F | CTTGACATGAATACAGCAGAGGAT | DQc、TQc |
| Cluster-0.121834-R | GCACCATCAATTTCAGCAACTT | |
| Cluster-0.121871-F | TCTGTGTCCTGCTCTTTGAAG | DQc、TQc |
| Cluster-0.121871-R | GCAACCTCAGAGATGGGAAG | |
| Cluster-0.121919-F | TAACCATAACGCCTCCATCTCT | DQc、TQc |
| Cluster-0.121919-R | TCATCTCACGAGACACAATAGACT | |
| Cluster-0.121949-F | TAACCATAACGCCTCCTTCTCT | DQc、TQc |
| Cluster-0.121949-R | GCTCCTACAATCACCACATCAG | |
| Cluster-0.155662-F | AGACTATGACGAGCAAGACAACT | DQc、TQc |
| Cluster-0.155662-R | GTTACCACACCACCGACTCT | |
| Cluster-0.119990-F | CTGTTGCCGTAACTTCACTGT | TQc |
| Cluster-0.119990-R | CTGAGAGATGGGAAGCGTTTC | |
| Cluster-0.120915-F | ATCATTCTCCAGGCATTTCTCAG | TQc |
| Cluster-0.120915-R | GGAACAGCCACAGCTTGTAT | |
| Cluster-0.121783-F | CAGGAAGTGTCAGCAACATCAA | TQc |
| Cluster-0.121783-R | CTGTTCTTCGCAAGGTTCATCT | |
| Cluster-0.121896-F | AGGAGTGAAGTGGTTGAAGAGTA | DQc |
| Cluster-0.121896-R | ACAATCATCCCAGGCACAATC | |
| Cluster-0.122437-F | TAACCATAACGCCTCCATCTCT | DQc |
| Cluster-0.122437-R | ATCAGTGTCCGCATGTGTAATC | |
| Cluster-0.122522-F | CCTCCATCTCTGCTTCTTCCT | DQc |
| Cluster-0.122522-R | TGTACCTGCGTGTCATCTCA | |
| 18S rRNA-F(内参基因) | CCTGAGAAACGGCTACCACA | DQc、TQc |
| 18S rRNA-R(内参基因) | CACCAGACTT GCCCTCCA |
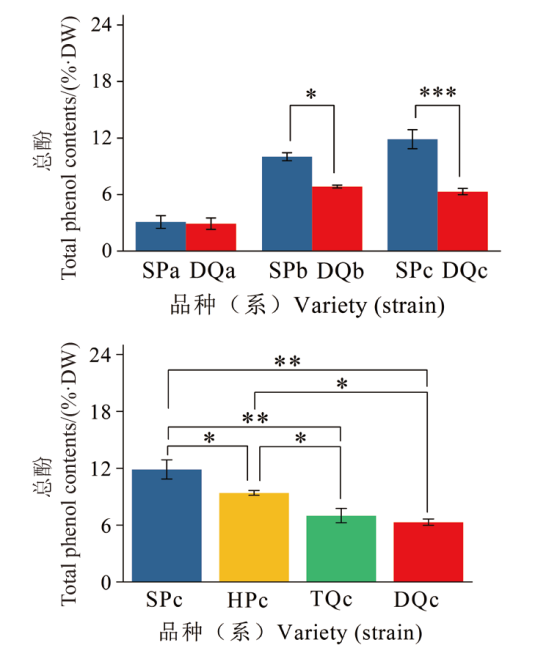
图1 橄榄果实成熟过程及成熟期总酚含量 *表示0.05水平差异显著,**表示0.01水平差异显著,***表示0.001水平差异显著
Fig. 1 Total phenol content in Chinese olive fruit ripening process and maturity period * indicates significant difference at 0.05 level, ** indicates significant difference at 0.01 level, and *** indicates significant difference at 0.001 level
| 类型Type | 序列条数Number of sequences | 序列平均长度Mean length/bp | N50/bp | N90/bp | 序列总碱基Total bases/bp |
|---|---|---|---|---|---|
| Unigene | 296 314 | 1 163 | 1 764 | 522 | 344 722 448 |
表2 橄榄转录组序列拼接结果
Table 2 Sequence splicing results of Chinese olive transcriptome
| 类型Type | 序列条数Number of sequences | 序列平均长度Mean length/bp | N50/bp | N90/bp | 序列总碱基Total bases/bp |
|---|---|---|---|---|---|
| Unigene | 296 314 | 1 163 | 1 764 | 522 | 344 722 448 |
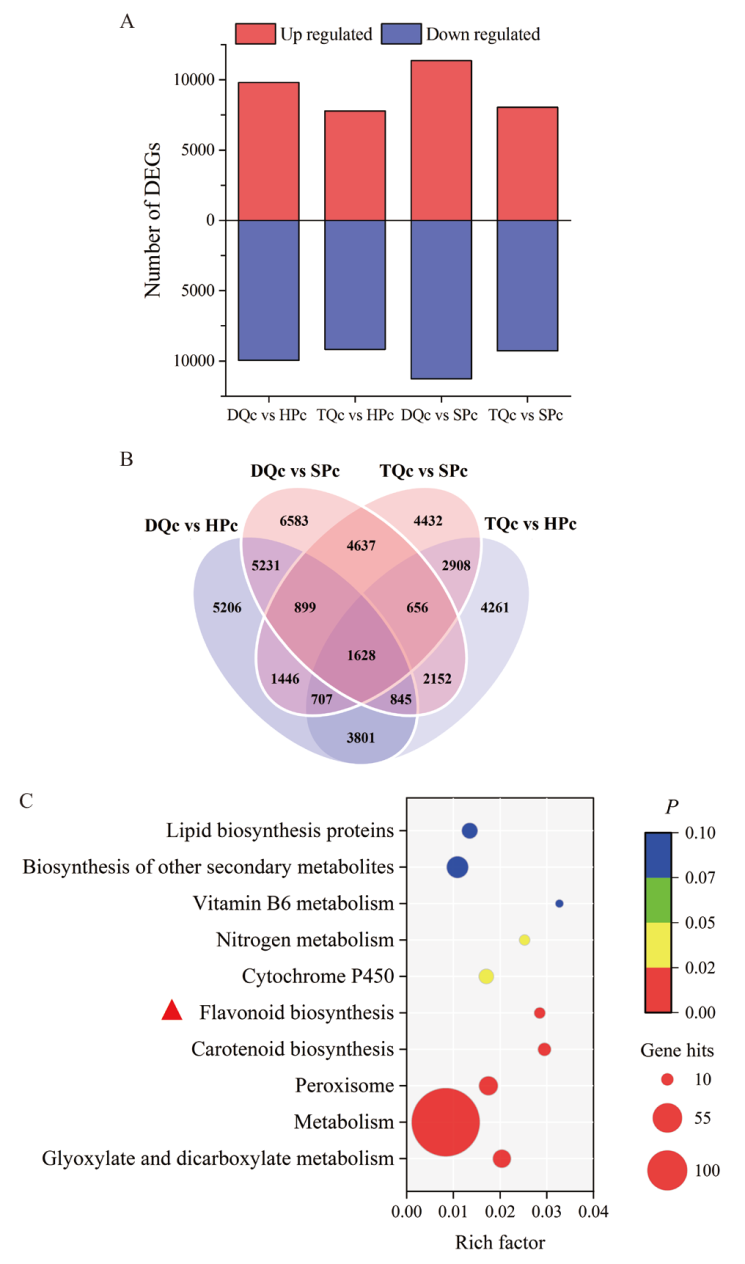
图4 不同品种(系)橄榄差异基因与KEGG富集分析 A:上下调差异基因,基因上/下调表达均是指低酚橄榄(DQc、TQc)相对于高酚橄榄(SPc、HPc);B:共有或独特差异基因;C:共有差异基因KEGG富集分析
Fig. 4 Differential genes and KEGG enrichment analysis of Chinese olive in different varieties(lines) A : Up-regulated and down-regulated differential genes, gene up/down-regulated expression refers to low phenolic Chinese olive(DQc, TQc)relative to high phenolic Chinese olive(SPc, HPc); B : common or unique differential genes ;C : KEGG enrichment analysis of common differential genes

图5 不同品种(系)橄榄成熟过程差异基因与KEGG富集分析 A:‘子阳1号’(SP)与‘东山长穗’(DQ)成熟过程差异基因,B:不同成熟时期差异基因的韦恩图,C:差异基因KEGG富集分析
Fig. 5 Differential genes and KEGG enrichment analysis of different varieties(lines)of Chinese olive ripening process A: Differential genes in the maturation process of ‘Ziyang 1’ (SP)and ‘Dongshanchangsui’ (DQ). B : Venn of differential genes in different maturation periods. C : KEGG enrichment analysis of differential genes
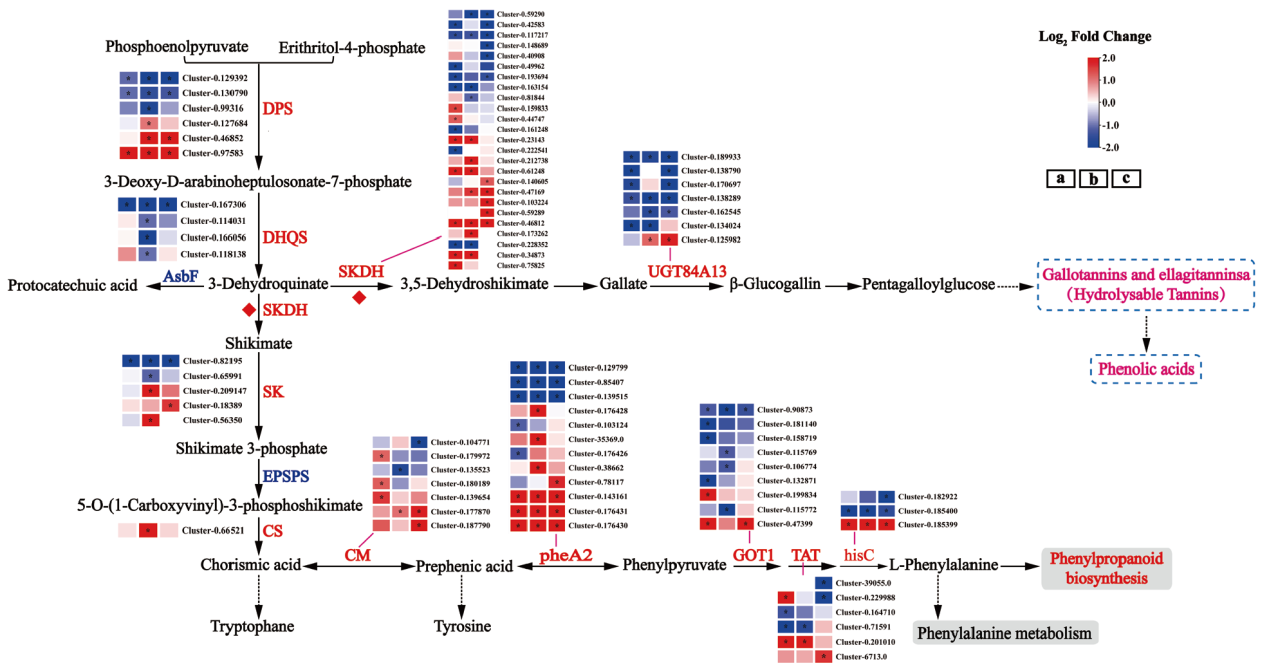
图6 不同品种(系)橄榄成熟过程莽草酸-水解单宁生物合成途径差异基因分析 DPS:3-脱氧-D-阿拉伯庚酮糖酸-7-磷酸合酶,DHQS:3-脱氢奎尼酸合酶,SKDH:莽草酸脱氢酶,UGTs:糖基转移酶,SK:莽草酸激酶,EPSPS:5-烯醇丙酮酰莽草酸-3-磷酸合酶,CS:分支酸合酶,CM:分支酸变位酶,pheA2:预苯酸脱水酶,GOT1:天冬氨酸氨基转移酶,TAT:酪氨酸氨基转移酶,hisC:磷酸组氨酸氨基转移酶;红色或蓝色的颜色块表示Log2FC(DQ/SP)的值,红色和蓝色分别表示低酚橄榄(DQ)相较于高酚橄榄(SP)上调和下调的基因,*表示P.adjust<0.05,下同
Fig. 6 Differential gene analysis of shikimic acid and hydrolyzed tannin biosynthesis pathway in different varieties(lines)of Chinese olive during maturation DPS : 3-deoxy-D-arabinoheptulose-7-phosphate synthase; DHQS : 3-dehydroquinic acid synthase; SKDH : shikimate dehydrogenase; UGTs : glycosyltransferase; SK : shikimate kinase; EPSPS : 5-enolpyruvylshikimate-3-phosphate synthase; CS : chorismate synthase; CM : chorismate mutase; pheA2 : prebenzoic acid dehydratase; GOT1 : aspartate aminotransferase; TAT : tyrosine aminotransferase; hisC : phosphohistidine aminotransferase ; Red or blue color blocks represent the value of Log2FC(DQ / SP), red and blue represent the up-regulated and down-regulated genes of low-phenol olive(DQ)compared to high-phenol olive(SP), respectively, and * indicates P.adjust < 0.05, the same below

图7 不同品种(系)橄榄成熟过程苯丙烷-类黄酮生物合成途径差异基因分析 PAL:苯丙氨酸解氨酶,C4H:肉桂酸-4-羟化酶,4CL:4-香豆酰辅酶A连接酶,CHS:查尔酮合成酶,CHI:查尔酮异构酶,IFS:异黄酮合酶,FNS:黄酮合酶,F3H:黄烷酮-3-羟化酶,F3'H:类黄酮 3'-羟化酶,F3'5'H:类黄酮 3'5'-羟化酶,FLS:黄酮醇合成酶,DFR:二氢黄酮醇还原酶,LAR:无色花色素还原酶,ANR:花青素还原酶;ANS:花青素合成酶
Fig. 7 Differential gene analysis in phenylpropanes and flavonoids biosynthesis pathways in different varieties(lines)of Chinese olive during maturation PAL : Phenylalanine ammonia lyase; C4H : cinnamic acid-4-hydroxylase; 4CL : 4-coumaroyl-CoA ligase; CHS : chalcone synthase; CHI : chalcone isomerase; IFS : isoflavone synthase; FNS : flavonoid synthase; F3H : flavanone-3-hydroxylase; F3'H : flavonoid 3'-hydroxylase; F3' 5'H : flavonoid 3' 5' -hydroxylase; FLS : flavonol synthase; DFR : dihydroflavonol reductase; LAR : achromatic anthocyanin reductase; ANR : anthocyanin reductase ; aNS : anthocyanin synthase
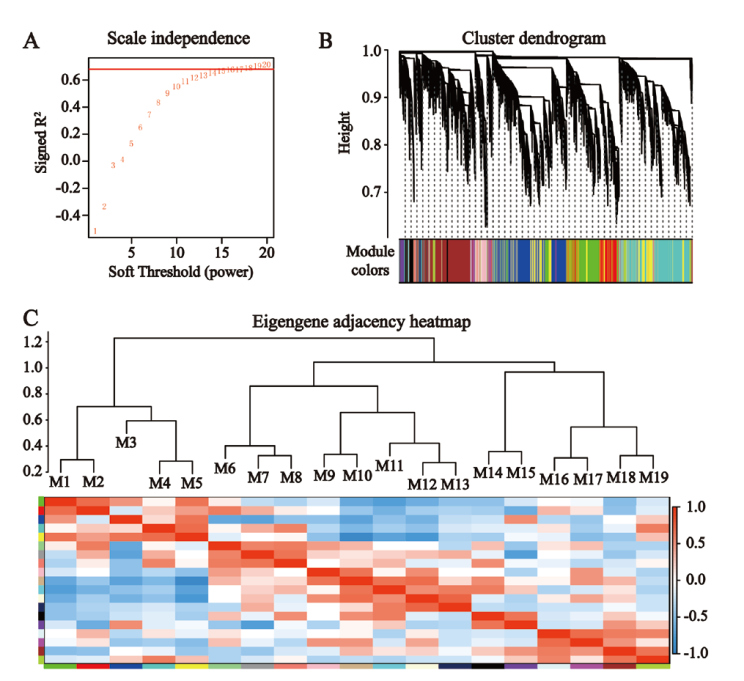
图8 基因共表达网络的可视化 A:确定最佳软阈值,B:共表达模块的集群树状图,C:共表达模块之间的相关性
Fig. 8 Visualization of gene co-expression network A: Determination of the optimal soft threshold. B: Cluster dendrogram of co-expression modules. C: Correlation between co-expression modules

图9 酚类化合物模块鉴定及其相关转录因子挖掘 A:模块与总酚相关性热图,每一行对应一个模块,每个格子的值分别表示相关系数R和P值;格子颜色表示相关性,红色表示正相关,蓝色表示负相关;B:酚类物质合成基因与转录因子的共表达网络分析,颜色越深表示基因在互作网络中越重要;C:酚类物质合成相关转录因子类别统计
Fig. 9 Module identification of phenolic compounds and mining of their associated transcription factors A:Module and trait correlation heat map,each row corresponds to a module. The values of each lattice indicate the correlation coefficients R and P, respectively. The lattice color indicates correlation, red indicates positive correlation, and blue indicates negative correlation. B:Co-expression network analysis of phenolic synthesis genes and transcription factors. Darker colors indicate that the gene is more important in the interaction network. C:Category statistics of transcription factors related to phenol synthesis
| [1] | 陈杰忠. 果树栽培学各论: 南方本[M]. 4版. 北京: 中国农业出版社, 2011: 139-145. |
| Chen JZ. Various theories on fruit tree cultivation: southern edition[M]. 4th ed. Beijing: China Agriculture Press, 2011: 139-145. | |
| [2] | 谢倩, 李易易, 张诗艳, 等. 基于模糊数学感官评价、理化特性与电子舌的橄榄鲜食品质分析[J]. 食品科学, 2023, 44(3): 69-78. |
|
Xie Q, Li YY, Zhang SY, et al. Quality analysis of table Cauarium album L. based on fuzzy mathematics sensory evaluation, physicochemical properties and electronic tongue[J]. Food Sci, 2023, 44(3): 69-78.
doi: 10.1111/jfds.1979.44.issue-1 URL |
|
| [3] | 林玉芳, 杜正花, 陈清西. 橄榄果实品质评价因子的筛选及指标确定[J]. 热带作物学报, 2014, 35(4): 805-810. |
| Lin YF, Du ZH, Chen QX. Selection of quality evaluation indices for Chinese olive[J]. Chin J Trop Crops, 2014, 35(4): 805-810. | |
| [4] | 池毓斌, 朱丽娟, 黄敏杰, 等. 鲜食橄榄品质综合评价模型的建立与验证[J]. 果树学报, 2017, 34(8): 1051-1060. |
| Chi YB, Zhu LJ, Huang MJ, et al. Establishment and verification of a comprehensive evaluation model for quality of fresh Chinese olive[J]. J Fruit Sci, 2017, 34(8): 1051-1060. | |
| [5] | 谢倩, 张诗艳, 叶清华, 等. 鲜食橄榄发育成熟过程中多酚及相关酶活性的动态变化[J]. 果树学报, 2019, 36(6): 774-784. |
| Xie Q, Zhang SY, Ye QH, et al. Dynamic changes of polyphenols and related enzymes activity during the development and maturation of Chinese olive(Canarium album L.)[J]. J Fruit Sci, 2019, 36(6): 774-784. | |
| [6] | 池毓斌. 橄榄果实品质特性及其代谢组学的初步研究[D]. 福州: 福建农林大学, 2017. |
| Chi YB. A preliminary study on the fruit quality characteristics and metabolism of Canarium album[D]. Fuzhou: Fujian Agriculture and Forestry University, 2017. | |
| [7] | 李泽坤. 橄榄(Canarium album(lour.)raeusch)果实成熟发育蔗糖代谢变化研究[D]. 福州: 福建农林大学, 2016. |
| Li ZK. Studyies of sucrose metabolism changes in Chinese olive fruit mature development[D]. Fuzhou: Fujian Agriculture and Forestry University, 2016. | |
| [8] |
彭真汾, 叶清华, 王威, 等. 普通橄榄和清橄榄果实游离氨基酸差异成分与谷氨酰胺代谢[J]. 食品科学, 2019, 40(4): 229-236.
doi: 10.7506/spkx1002-6630-20171228-354 |
| Peng ZF, Ye QH, Wang W, et al. Differences in free amino acid composition of fruits of common olive and sweet olive and their glutamine metabolism characteristics[J]. Food Sci, 2019, 40(4): 229-236. | |
| [9] | 常强. 橄榄果实酚类物质及其抗氧化活性研究[D]. 福州: 福建农林大学, 2017. |
| Chang Q. Investigation of polyphenol compounds and their antioxidant activity in the fruits of Chinese olive[D]. Fuzhou: Fujian Agriculture and Forestry University, 2017. | |
| [10] | 谢倩. 橄榄(Canarium album(Lour.)Rauesch.)果实发育成熟过程多酚及相关酶活性研究[D]. 福州: 福建农林大学, 2014. |
| Xie Q. Polyphenol components and related enzyme activities during the Chinese olive(Canarium album(Lour.)Rauesch.)fruit development and ripening[D]. Fuzhou: Fujian Agriculture and Forestry University, 2014. | |
| [11] | 林玉芳. 福建橄榄(Canarium album(lour.)raeusch.)若干功能成分和品质相关指标的研究[D]. 福州: 福建农林大学, 2012. |
| Lin YF. Studies on some functional components and quality indexes for Chinese olive fruits in Fujian Province[D]. Fuzhou: Fujian Agriculture and Forestry University, 2012. | |
| [12] | 何志勇, 夏文水. 橄榄果实中酚类化合物的分析研究[J]. 安徽农业科学, 2008, 36(26): 11406-11407. |
| He ZY, Xia WS. Analysis of phenolic compounds in Canarium album(Lour.)raeusch fruit[J]. J Anhui Agric Sci, 2008, 36(26): 11406-11407. | |
| [13] |
蔡净蓉, 王杰, 赵俊跃, 等. 成熟期不同橄榄品种(系)果实代谢组及其差异[J]. 热带作物学报, 2022, 43(11): 2304-2315.
doi: 10.3969/j.issn.1000-2561.2022.11.015 |
| Cai JR, Wang J, Zhao JY, et al. Metabolomics and its difference of Chinese olive fruit of different varieties(lines)during the ripening period[J]. Chin J Trop Crops, 2022, 43(11): 2304-2315. | |
| [14] |
侯黔东, 沈天娇, 余欢欢, 等. 甜樱桃GH3基因家族全基因组鉴定与表达分析[J]. 园艺学报, 2021, 48(12): 2360-2374.
doi: 10.16420/j.issn.0513-353x.2020-0911 |
| Hou QD, Shen TJ, Yu HH, et al. Genome-wide identification and expression analysis of Prunus avium gretchen Hagen 3(GH3)gene family[J]. Acta Hortic Sin, 2021, 48(12): 2360-2374. | |
| [15] |
Broun P. Transcription factors as tools for metabolic engineering in plants[J]. Curr Opin Plant Biol, 2004, 7(2): 202-209.
doi: 10.1016/j.pbi.2004.01.013 pmid: 15003222 |
| [16] |
Vom Endt D, Kijne JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators?[J]. Phytochemistry, 2002, 61(2): 107-114.
pmid: 12169302 |
| [17] |
Yang CQ, Fang X, Wu XM, et al. Transcriptional regulation of plant secondary metabolism[J]. J Integr Plant Biol, 2012, 54(10): 703-712.
doi: 10.1111/jipb.2012.54.issue-10 URL |
| [18] | 谭西北, 李鹏, 孙磊, 等. 基于WGCNA的刺葡萄抗白腐病关键基因的发掘[J]. 果树学报, 2023, 40(4): 653-668. |
| Tan XB, Li P, Sun L, et al. Discovery of key genes for White Rot resistance in Vitis davidii based on WGCNA[J]. J Fruit Sci, 2023, 40(4): 653-668. | |
| [19] |
李贵生. 猕猴桃‘金艳’和‘红阳’果实转录组的比较分析[J]. 园艺学报, 2021, 48(6): 1183-1196.
doi: 10.16420/j.issn.0513-353x.2020-0510 |
| Li GS. Comparative analysis of‘Jinyan’ and‘Hongyang’ kiwifruit transcriptomes[J]. Acta Hortic Sin, 2021, 48(6): 1183-1196. | |
| [20] |
辛建攀, 李燕, 赵楚, 等. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1151 |
| Xin JP, Li Y, Zhao C, et al. Transcriptome sequencing in the leaves of Pontederia cordata with cadmium exposure and gene mining in phenypropanoid pathways[J]. Biotechnol Bull, 2022, 38(6): 198-210. | |
| [21] | 赵权. 葡萄酚类物质及其生物合成相关结构基因表达[D]. 哈尔滨: 东北林业大学, 2010. |
| Zhao Q. Phenolic compounds and expression of related biosynthesis structural genes in grapevine[D]. Harbin: Northeast Forestry University, 2010. | |
| [22] | 林玉芳, 陈清西, 关夏玉, 等. 橄榄总多酚提取工艺优化研究[J]. 中国农学通报, 2011, 27(5): 396-400. |
| Lin YF, Chen QX, Guan XY, et al. Extraction of total polyphenol from Chinese olive(Canarium ablum L.)[J]. Chin Agric Sci Bull, 2011, 27(5): 396-400. | |
| [23] |
谢倩, 王威, 陈清西. 橄榄多酚含量测定方法的比较[J]. 食品科学, 2014, 35(8): 204-207.
doi: 10.7506/spkx1002-6630-201408040 |
| Xie Q, Wang W, Chen QX. Comparative study on three different methods for the determination of total phenolics in Chinese olive[J]. Food Sci, 2014, 35(8): 204-207. | |
| [24] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [25] |
Wang YR, Suo YJ, Han WJ, et al. Comparative transcriptomic and metabolomic analyses reveal differences in flavonoid biosynthesis between PCNA and PCA persimmon fruit[J]. Front Plant Sci, 2023, 14: 1130047.
doi: 10.3389/fpls.2023.1130047 URL |
| [26] |
Liu HX, Liu Q, Chen YL, et al. Full-length transcriptome sequencing provides insights into flavonoid biosynthesis in Camellia nitidissima Petals[J]. Gene, 2023, 850: 146924.
doi: 10.1016/j.gene.2022.146924 URL |
| [27] |
Ye QH, Zhang SY, Qiu NN, et al. Identification and characterization of glucosyltransferase that forms 1-galloyl- β-d-glucogallin in Camellia nitidissima L., a functional fruit rich in hydrolysable tannins[J]. Molecules, 2021, 26(15): 4650.
doi: 10.3390/molecules26154650 URL |
| [28] |
Liu WX, Feng Y, Yu SH, et al. The flavonoid biosynthesis network in plants[J]. Int J Mol Sci, 2021, 22(23): 12824.
doi: 10.3390/ijms222312824 URL |
| [29] | 李小溪. 葡萄果实莽草酸途径和类黄酮代谢协同调节机制的研究[D]. 北京: 中国农业大学, 2016. |
| Li XX. Collaborative expression mechanism between shikimate pathway and flavonoid metabolism[D]. Beijing: China Agricultural University, 2016. | |
| [30] |
Routaboul JM, Kerhoas L, Debeaujon I, et al. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana[J]. Planta, 2006, 224(1): 96-107.
doi: 10.1007/s00425-005-0197-5 URL |
| [31] |
Saito K, Yonekura-Sakakibara K, Nakabayashi R, et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity[J]. Plant Physiol Biochem, 2013, 72: 21-34.
doi: 10.1016/j.plaphy.2013.02.001 URL |
| [32] | Chen CX, Li AL. Transcriptome analysis of differentially expressed genes involved in proanthocyanidin accumulation in the rhizomes of Fagopyrum dibotrys and an irradiation-induced mutant[J]. Front Physiol, 2016, 7: 100. |
| [33] | 张小郁, 李文广, 高明堂, 等. 葡萄籽中原花青素对心肌细胞的保护作用[J]. 中药药理与临床, 2001, 17(6): 14-16. |
| Zhang XY, Li WG, Gao MT, et al. Protective effect on injured myocardial cells of proanthoc yanidins in vitro[J]. Pharmacol Clin Chin Mater Med, 2001, 17(6): 14-16. | |
| [34] |
Chen JH, Hou N, Xv X, et al. Flavonoid synthesis and metabolism during the fruit development in hickory(Carya cathayensis)[J]. Front Plant Sci, 2022, 13: 896421.
doi: 10.3389/fpls.2022.896421 URL |
| [35] |
Xia X, Gong R, Zhang CY. Integrative analysis of transcriptome and metabolome reveals flavonoid biosynthesis regulation in Rhododendron pulchrum petals[J]. BMC Plant Biol, 2022, 22(1): 401.
doi: 10.1186/s12870-022-03762-y pmid: 35974307 |
| [36] | 陈为凯. 一年两收栽培模式下葡萄果实靶向代谢组和转录组研究[D]. 北京: 中国农业大学, 2018. |
| Chen WK. Study of targeted metabolome and transcriptome in grape berries grown under double cropping viticulture system[D]. Beijing: China Agricultural University, 2018. | |
| [37] |
Jeong ST, Goto-Yamamoto N, Hashizume K, et al. Expression of the flavonoid 3'-hydroxylase and flavonoid 3', 5'-hydroxylase genes and flavonoid composition in grape(Vitis vinifera)[J]. Plant Sci, 2006, 170(1): 61-69.
doi: 10.1016/j.plantsci.2005.07.025 URL |
| [38] |
Springob K, Nakajima JI, Yamazaki M, et al. Recent advances in the biosynthesis and accumulation of anthocyanins[J]. Nat Prod Rep, 2003, 20(3): 288-303.
doi: 10.1039/b109542k pmid: 12828368 |
| [39] | 李小兰, 张明生, 吕享. 植物花青素合成酶ANS基因的研究进展[J]. 植物生理学报, 2016, 52(6): 817-827. |
| Li XL, Zhang MS, Lü X. The research progress on plant anthocyanin synthetase ANS gene[J]. Plant Physiol J, 2016, 52(6): 817-827. | |
| [40] |
Bogs J, Downey MO, Harvey JS, et al. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves[J]. Plant Physiol, 2005, 139(2): 652-663.
doi: 10.1104/pp.105.064238 pmid: 16169968 |
| [41] |
Yamasaki K, Kigawa T, Seki M, et al. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution[J]. Trends Plant Sci, 2013, 18(5): 267-276.
doi: 10.1016/j.tplants.2012.09.001 pmid: 23040085 |
| [42] | 沈德绪. 果树育种学: 全国高等农业院校教材果树专业园艺专业用[M]. 第2版. 北京: 中国农业出版社, 2000. |
| Shen DX. Fruit tree breeding: textbook for fruit tree specialty and horticulture specialty in national agricultural colleges and universities[M]. 2th ed. Beijing: China Agriculture Press, 2000. | |
| [43] | 朱军. 遗传学[M]. 4版. 北京: 中国农业出版社, 2018. |
| Zhu J. Genetics[M]. 4th ed. Beijing: China Agriculture Press, 2018. | |
| [44] | 穆红梅, 杜秀菊, 张秀省, 等. 植物MYB转录因子调控苯丙烷类生物合成研究[J]. 北方园艺, 2015(24): 171-174. |
| Mu HM, Du XJ, Zhang XS, et al. Study on plants MYB transcription factors regulate biological synthesis of phenylpropanoid metabolism[J]. North Hortic, 2015(24): 171-174. | |
| [45] |
Hichri I, Barrieu F, Bogs J, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway[J]. J Exp Bot, 2011, 62(8): 2465-2483.
doi: 10.1093/jxb/erq442 pmid: 21278228 |
| [46] |
Yoshida K, Ma DW, Constabel CP. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes[J]. Plant Physiol, 2015, 167(3): 693-710.
doi: 10.1104/pp.114.253674 pmid: 25624398 |
| [47] |
Premathilake AT, Ni JB, Bai SL, et al. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit[J]. Planta, 2020, 252(4): 59.
doi: 10.1007/s00425-020-03473-4 |
| [48] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [49] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [50] |
孙庆国, 姜生辉, 房鸿成, 等. 苹果MdNAC9的克隆及其调控黄酮醇合成功能的鉴定[J]. 园艺学报, 2019, 46(11): 2073-2081.
doi: 10.16420/j.issn.0513-353x.2018-1070 |
| Sun QG, Jiang SH, Fang HC, et al. Cloning of MdNAC9 and functional of its regulation on flavonol synthesis[J]. Acta Hortic Sin, 2019, 46(11): 2073-2081. | |
| [51] |
Han H, Xu F, Li YT, et al. Genome-wide characterization of bZIP gene family identifies potential members involved in flavonoids biosynthesis in Ginkgo biloba L[J]. Sci Rep, 2021, 11(1): 23420.
doi: 10.1038/s41598-021-02839-2 |
| [52] |
Liu WJ, Wang YC, Yu L, et al. MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5[J]. J Agric Food Chem, 2019, 67(32): 8783-8793.
doi: 10.1021/acs.jafc.9b02920 URL |
| [53] |
Mao ZL, Jiang HY, Wang S, et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple[J]. Plant Sci, 2021, 306: 110848.
doi: 10.1016/j.plantsci.2021.110848 URL |
| [54] |
An JP, Qu FJ, Yao JF, et al. Erratum: the bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple[J]. Hortic Res, 2017, 4: 17056.
doi: 10.1038/hortres.2017.56 URL |
| [55] | 罗曼. C2H2型锌指蛋白DkZF6在柿原花青素生物合成中的功能解析[D]. 武汉: 华中农业大学, 2021. |
| Luo M. Functional characterization of C2H2 type zinc finger DkZF6 in persimmon proanthocyanidins biosynthesis[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [56] |
Jamil W, Wu W, Gong H, et al. C2H2-type zinc finger proteins(DkZF1/2)synergistically control persimmon fruit deastringency[J]. Int J Mol Sci, 2019, 20(22): 5611.
doi: 10.3390/ijms20225611 URL |
| [57] | 黄萧天. 水稻Lec/B3基因调控种子休眠的作用机制的研究[D]. 自贡: 四川轻化工大学, 2021. |
| Huang XT. The mechanism of Lec/B3 genes in determination of seed dormancy in rice[D]. Zigong: Sichuan University of Science & Engineering, 2021. |
| [1] | 陈艳梅. 蛋白质翻译后修饰之间的互作关系及其协同调控机理[J]. 生物技术通报, 2024, 40(2): 1-8. |
| [2] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [3] | 路喻丹, 刘晓驰, 冯新, 陈桂信, 陈义挺. 猕猴桃BBX基因家族成员鉴定与转录特征分析[J]. 生物技术通报, 2024, 40(2): 172-182. |
| [4] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [5] | 周会汶, 吴兰花, 韩德鹏, 郑伟, 余跑兰, 吴杨, 肖小军. 甘蓝型油菜种子硫苷含量全基因组关联分析[J]. 生物技术通报, 2024, 40(1): 222-230. |
| [6] | 吴圳, 张明英, 闫锋, 李依民, 高静, 颜永刚, 张岗. 掌叶大黄(Rheum palmatum L.)WRKY基因家族鉴定与分析[J]. 生物技术通报, 2024, 40(1): 250-261. |
| [7] | 王斌, 袁晓, 蒋园园, 王玉昆, 肖艳辉, 何金明. bHLH96的克隆及其在薄荷萜烯生物合成调控中的功能[J]. 生物技术通报, 2024, 40(1): 281-293. |
| [8] | 陈治民, 李翠, 韦继天, 李昕然, 刘峄, 郭强. 绿原酸生物合成调控及其应用研究进展[J]. 生物技术通报, 2024, 40(1): 57-71. |
| [9] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [10] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [11] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [12] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [13] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [14] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [15] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||