








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 290-299.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0979
王周1( ), 余杰1, 王金华1,2,3, 王永泽1,2,3, 赵筱1,2,3(
), 余杰1, 王金华1,2,3, 王永泽1,2,3, 赵筱1,2,3( )
)
收稿日期:2023-10-19
出版日期:2024-05-26
发布日期:2024-05-23
通讯作者:
赵筱,女,博士,研究方向:微生物改造及发酵;E-mail: zhaoxiao1@hbut.edu.cn作者简介:王周,男,硕士研究生,研究方向:生物工程;E-mail: 2797807952@qq.com
基金资助:
WANG Zhou1( ), YU Jie1, WANG Jin-hua1,2,3, WANG Yong-ze1,2,3, ZHAO Xiao1,2,3(
), YU Jie1, WANG Jin-hua1,2,3, WANG Yong-ze1,2,3, ZHAO Xiao1,2,3( )
)
Received:2023-10-19
Published:2024-05-26
Online:2024-05-23
摘要:
【目的】为解决在D-乳酸工业发酵生产中使用农业粗加工或废弃物等廉价原料(含有少量L-乳酸)而导致终产物D-乳酸光纯降低的问题。【方法】构建了带有不同启动子的L-乳酸脱氢酶基因lldD的表达质粒:pUC19-PLD(PpflBp6)、pUC19-NLD(PnirB)及pUC19-PNLD(PpflBp6-PnirB),并将它们分别转化入大肠杆菌D-乳酸工程菌HBUT-D中,得到菌株HBUT-D3、HBUT-D5和HBUT-D7。通过LldD的酶活检测以及对NBS培养基(添加1 g/L L-乳酸)发酵结果的TOPSIS多元评估,优选出可快速去除L-乳酸且不影响D-乳酸发酵的菌株,并使用农业廉价原料进行发酵。【结果】HBUT-D7的LldD比酶活为64 U/g,L -乳酸的消耗速率为34 mg/(L·h),D-乳酸的生产强度为4.09 g/(L·h),综合评估为最优菌株。以玉米浆为原料进行发酵时,HBUT-D及HBUT-D7的L-乳酸消耗速率分别为10.41 mg/(L·h)及34.75 mg/(L·h);D-乳酸生产强度分别为4.24 g/(L·h)及3.87 g/(L·h);D-乳酸光学纯度分别为99.07%及99.92%。以糖蜜为原料进行发酵时,HBUT-D及HBUT-D7的L-乳酸消耗速率为6.87 mg/(L·h)和17.18 mg/(L·h);D-乳酸生产强度分别为1.93 g/(L·h)及1.88 g/(L·h);D-乳酸光学纯度分别为99.22%及99.99%。【结论】厌氧诱导启动子的表达质粒可提高菌株HBUT-D7的L-乳酸脱氢酶酶活,使其在使用廉价原料发酵时能有效消除L-乳酸,提高发酵终产物D-乳酸的光学纯度。
王周, 余杰, 王金华, 王永泽, 赵筱. 厌氧表达乳酸脱氢酶以提高大肠杆菌产D-乳酸光学纯度[J]. 生物技术通报, 2024, 40(5): 290-299.
WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli[J]. Biotechnology Bulletin, 2024, 40(5): 290-299.
| 菌株及质粒 Strain and plasmid | 特征 Characteristic | 来源 Source |
|---|---|---|
| HBUT-D | W ∆frdBC ∆pta ∆adhE ∆pflB ∆aldA | 本实验室保藏 |
| HBUT-DP | HBUT-D carrying pUC19 | 本研究 |
| HBUT-D3 | HBUT-D carrying pUC19-PLD | 本研究 |
| HBUT-D5 | HBUT-D carrying pUC19-PNLD | 本研究 |
| HBUT-D7 | HBUT-D carrying pUC19-NLD | 本研究 |
| pUC19-PLD | pUC19::PpflBp6-lldD, Apr | 本研究 |
| pUC19-NLD | pUC19::PnirB-lldD, Apr | 本研究 |
| pUC19-PNLD | pUC19::PpflBp6-PnirB-lldD, Apr | 本研究 |
表1 本研究菌株及质粒
Table 1 Strains and plasmids in this study
| 菌株及质粒 Strain and plasmid | 特征 Characteristic | 来源 Source |
|---|---|---|
| HBUT-D | W ∆frdBC ∆pta ∆adhE ∆pflB ∆aldA | 本实验室保藏 |
| HBUT-DP | HBUT-D carrying pUC19 | 本研究 |
| HBUT-D3 | HBUT-D carrying pUC19-PLD | 本研究 |
| HBUT-D5 | HBUT-D carrying pUC19-PNLD | 本研究 |
| HBUT-D7 | HBUT-D carrying pUC19-NLD | 本研究 |
| pUC19-PLD | pUC19::PpflBp6-lldD, Apr | 本研究 |
| pUC19-NLD | pUC19::PnirB-lldD, Apr | 本研究 |
| pUC19-PNLD | pUC19::PpflBp6-PnirB-lldD, Apr | 本研究 |
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| P1 | CCGTGACTTAAGAAAATTTATAC |
| P2 | CTGGCTGCGGAAATAATCATTTTTGCCTCGATTTCTTTTC |
| P3 | GAAAAGAAATCGAGGCAAAAATGTCTTCCATGACAACAACTG |
| P4 | ACATACAGCGCCGAACGGTC |
| P5 | CAGCAATATACCCATTAAGGAGTAT |
| P6 | AACGACTATGCCGCATTC |
| P7 | TTCGCCTGATGCTCCC |
| P8 | CTGGCTGCGGAAATAATCATACTAACTCTCTCTTTATTAAGTCGGC |
| P9 | GCCGACTTAATAAAGAGAGAGTTAGTATGATTATTTCCGCAGCCAG |
| P10 | ATATGCGGTGTGAAATACC |
| P11 | GTTTTACAACGTCGTGACTTTGGATAATCAAATATTTACTCCGT |
| P12 | TAAGGAGAAAATACCGCATCTATGCCGCATTCCCTTT |
| P13 | AGTCACGACGTTGTAAAAC |
| P14 | ATGCGGTATTTTCTCCTTA |
| P15 | ATAGTTTAGCGGCCGCATTCTTATACAGATGCGTAAGGAGAAAA |
| P16 | ATAAGAATGCGGCCGCTAAACTATTTTTCTCCTTACGCATCTGT |
| P17 | GCTCATACCTGAATGCGCA |
| P18 | CGGAAGAGCGCCCAAT |
| P19 | CTTATCGCCACTGGCAGCC |
| P20 | TGTAGGTCGTTTCGCTCCAA |
表2 本研究所用引物
Table 2 Primers in this study
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| P1 | CCGTGACTTAAGAAAATTTATAC |
| P2 | CTGGCTGCGGAAATAATCATTTTTGCCTCGATTTCTTTTC |
| P3 | GAAAAGAAATCGAGGCAAAAATGTCTTCCATGACAACAACTG |
| P4 | ACATACAGCGCCGAACGGTC |
| P5 | CAGCAATATACCCATTAAGGAGTAT |
| P6 | AACGACTATGCCGCATTC |
| P7 | TTCGCCTGATGCTCCC |
| P8 | CTGGCTGCGGAAATAATCATACTAACTCTCTCTTTATTAAGTCGGC |
| P9 | GCCGACTTAATAAAGAGAGAGTTAGTATGATTATTTCCGCAGCCAG |
| P10 | ATATGCGGTGTGAAATACC |
| P11 | GTTTTACAACGTCGTGACTTTGGATAATCAAATATTTACTCCGT |
| P12 | TAAGGAGAAAATACCGCATCTATGCCGCATTCCCTTT |
| P13 | AGTCACGACGTTGTAAAAC |
| P14 | ATGCGGTATTTTCTCCTTA |
| P15 | ATAGTTTAGCGGCCGCATTCTTATACAGATGCGTAAGGAGAAAA |
| P16 | ATAAGAATGCGGCCGCTAAACTATTTTTCTCCTTACGCATCTGT |
| P17 | GCTCATACCTGAATGCGCA |
| P18 | CGGAAGAGCGCCCAAT |
| P19 | CTTATCGCCACTGGCAGCC |
| P20 | TGTAGGTCGTTTCGCTCCAA |
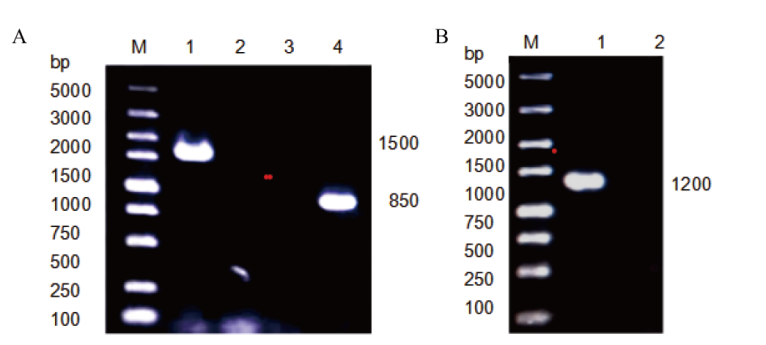
图1 不同质粒的验证 A:质粒pUC19-PLD及pUC19-NLD验证(M:5000 marker;1:pUC19-PLD;2-3:pUC19;4:pUC19-PNLD);B:质粒pUC19-PNLD验证(M:5000 marker;1:pUC19-NLD;2:pUC19)
Fig. 1 Validation of different plasmids A: Validation of plasmid pUC19-PLD and plasmid pUC19-NLD(M:5000 marker;1:pUC19-PLD;2-3:pUC19;4:pUC19-PNLD). B: Validation of plasmid pUC19-PNLD(M:5000 marker;1:pUC19-NLD;2:pUC19)
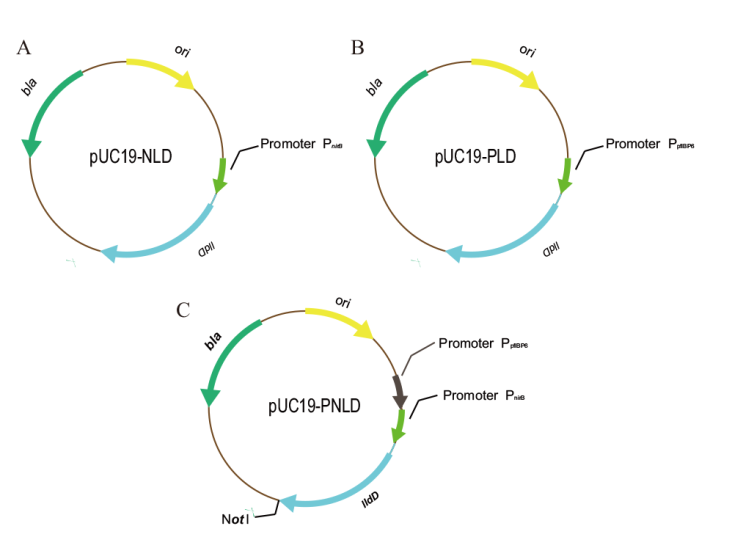
图2 不同启动子的lldD表达质粒 A:PnirB启动子重组质粒;B:PpflBP6启动子重组质粒;C:PpflBP6-PnirB串联启动子质粒
Fig. 2 Plasmids expressing lldD from different promoters A: PnirB promoter recombinant plasmid. B: PpflBP6 promoter recombinant plasmid. C: PpflBP6-PnirB tandem promoter plasmid
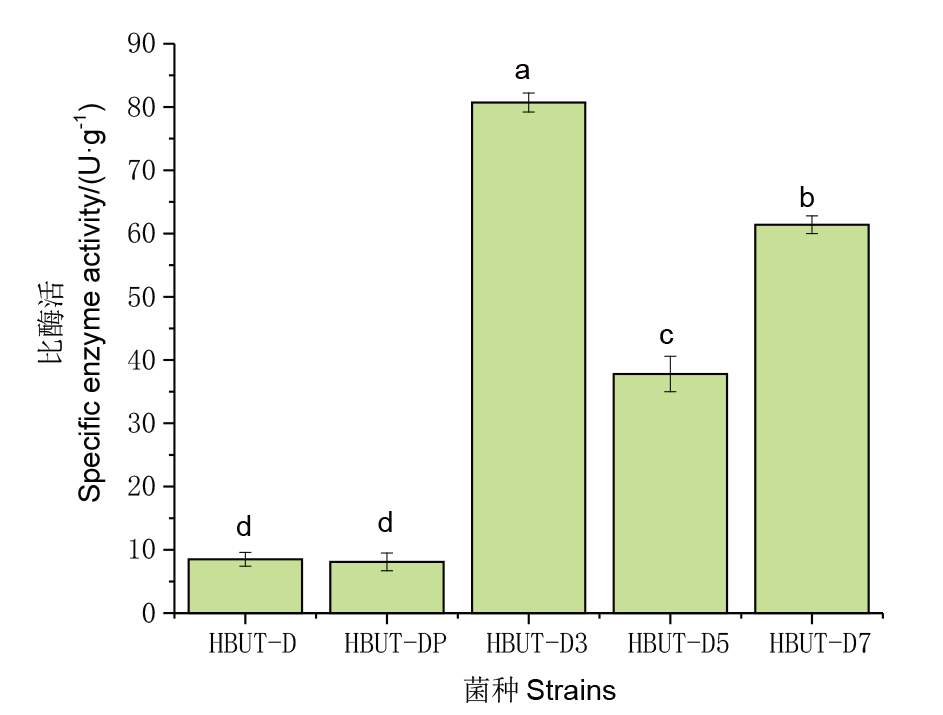
图3 不同菌株LldD比酶活 菌株HBUT-D为空白对照;菌株HBUT-DP为HBUT-D转入pUC19质粒;菌株HBUT-D3为HBUT-D转入质粒pUC19-PLD;菌株HBUT-D5为HBUT-D转入质粒pUC19-NLD;菌株HBUT-D7为HBUT-D转入质粒pUC19-PNLD。不同小写字母代表不同菌株的LldD比酶活的差异显著性(P<0.05)
Fig. 3 LldD specific enzyme activities from different strains Strain HBUT-D is the blank control, strain HBUT-DP was transferred to the plasmid pUC19 by HBUT-D, strain HBUT-D3 to plasmid pUC19-PLD, strain HBUT-D5 to plasmid pUC19-NLD by HBUT-D, and strain HBUT-D7 to plasmid pUC19-PNLD. Different lowercase letters indicate significant differences in LldD specific enzyme activities of different strains(P<0.05)
| 菌株 Strain | 酶活Enzyme activity/U | 蛋白量Protein content/mg | 比酶活Specific enzyme activity/(U·g-1) |
|---|---|---|---|
| HBUT-DP | 20±6 | 2 489 | 8.5±1.1 |
| HBUT-D | 20±4 | 2 596 | 8.1±1.4 |
| HBUT-D3 | 632±12 | 7 893 | 80.7±1.5 |
| HBUT-D5 | 275 ±10 | 7 274 | 37.8±2.8 |
| HBUT-D7 | 483±11 | 7 863 | 61.4±1.4 |
表3 不同菌株LldD酶活结果
Table 3 LldD enzyme activity results for different strains
| 菌株 Strain | 酶活Enzyme activity/U | 蛋白量Protein content/mg | 比酶活Specific enzyme activity/(U·g-1) |
|---|---|---|---|
| HBUT-DP | 20±6 | 2 489 | 8.5±1.1 |
| HBUT-D | 20±4 | 2 596 | 8.1±1.4 |
| HBUT-D3 | 632±12 | 7 893 | 80.7±1.5 |
| HBUT-D5 | 275 ±10 | 7 274 | 37.8±2.8 |
| HBUT-D7 | 483±11 | 7 863 | 61.4±1.4 |

图4 不同菌株无机盐培养基的发酵 A:不同菌株的L-乳酸消耗曲线;B:不同菌株的D-乳酸生产曲线;C:不同菌株葡萄糖消耗曲线;D:不同菌株生长曲线
Fig. 4 Fermentation of different strains in inorganic salt medium A: L-lactate consumption curves of different strains. B: Acid production curves of D-lactic acid of different strains. C: Glucose consumption curves of different strains. D: Growth curves of different strains
| 菌株 Strain | L-乳酸消耗速率Rate of L-lactate depletion/(mg·L-1·h-1) | L-乳酸消耗量L-lactate depletion concentration/(g·L-1) | D-乳酸生产强度D-lactate productivity/(g·L-1) | 葡萄糖消耗速率Rate of glucose consumption/(g·L-1·h-1) | OD600 nm | D-乳酸糖酸转化率D-lactate yield/% | 初始L-乳酸浓度Initial L -lactate concentration/(g·L-1) |
|---|---|---|---|---|---|---|---|
| HBUT-D | 8±0.13d | 0.21±0.01d | 4.09±0.11a | 4.16±0.04a | 3.22±0.01a | 98.31±0.21 | 1.01±0.11 |
| HBUT-D3 | 40±0.32a | 0.98±0.02a | 2.45±0.04c | 2.52±0.06d | 2.75±0.02b | 97.21±0.11 | 1.02±0.02 |
| HBUT-D5 | 14±0.33c | 0.34±0.01c | 4.09±0.04a | 4.15±0.08b | 3.12±0.01a | 98.55±0.33 | 1.04±0.02 |
| HBUT-D7 | 34±0.23b | 0.82±0.01b | 3.92±0.03b | 3.98±0.01c | 2.97±0.01a | 98.49±0.31 | 1.02±0.05 |
表4 不同菌株无机盐培养基发酵数据
Table 4 Data on the fermentation of different strains in inorganic salt medium
| 菌株 Strain | L-乳酸消耗速率Rate of L-lactate depletion/(mg·L-1·h-1) | L-乳酸消耗量L-lactate depletion concentration/(g·L-1) | D-乳酸生产强度D-lactate productivity/(g·L-1) | 葡萄糖消耗速率Rate of glucose consumption/(g·L-1·h-1) | OD600 nm | D-乳酸糖酸转化率D-lactate yield/% | 初始L-乳酸浓度Initial L -lactate concentration/(g·L-1) |
|---|---|---|---|---|---|---|---|
| HBUT-D | 8±0.13d | 0.21±0.01d | 4.09±0.11a | 4.16±0.04a | 3.22±0.01a | 98.31±0.21 | 1.01±0.11 |
| HBUT-D3 | 40±0.32a | 0.98±0.02a | 2.45±0.04c | 2.52±0.06d | 2.75±0.02b | 97.21±0.11 | 1.02±0.02 |
| HBUT-D5 | 14±0.33c | 0.34±0.01c | 4.09±0.04a | 4.15±0.08b | 3.12±0.01a | 98.55±0.33 | 1.04±0.02 |
| HBUT-D7 | 34±0.23b | 0.82±0.01b | 3.92±0.03b | 3.98±0.01c | 2.97±0.01a | 98.49±0.31 | 1.02±0.05 |
| 菌株 Strain | D+ | D- | C- | 排序Sort |
|---|---|---|---|---|
| HBUT-D | 0.334 3 | 0.139 7 | 0.294 8 | 4 |
| HBUT-D3 | 0.141 2 | 0.334 3 | 0.702 9 | 2 |
| HBUT-D5 | 0.271 6 | 0.154 2 | 0.362 2 | 3 |
| HBUT-D7 | 0.070 0 | 0.296 3 | 0.808 8 | 1 |
表5 各目标值与理想点之间的欧氏距离 D+和 D-以及相对距离C-
Table 5 Euclidean distance D+ and D- between each target value and the ideal point and the relative closeness C- of each target
| 菌株 Strain | D+ | D- | C- | 排序Sort |
|---|---|---|---|---|
| HBUT-D | 0.334 3 | 0.139 7 | 0.294 8 | 4 |
| HBUT-D3 | 0.141 2 | 0.334 3 | 0.702 9 | 2 |
| HBUT-D5 | 0.271 6 | 0.154 2 | 0.362 2 | 3 |
| HBUT-D7 | 0.070 0 | 0.296 3 | 0.808 8 | 1 |
| 菌株 Strain | D-乳酸糖酸转化率D-lactate yield/% | D-乳酸光纯D-lactate optical purty/% | D-乳酸生产强度D-lactate productivity/(g·L-1·h-1) | L-乳酸消耗速率Rate of L-lac consumption/(mg·L-1·h-1) | 初始L-乳酸浓度Initial L-lac concentration/(g·L-1) |
|---|---|---|---|---|---|
| HBUT-D | 97.64±0.01 | 99.22±0.02 | 4.24±0.04 | 10.41±0.01*** | 1.0±0.02 |
| HBUT-D7 | 96.89±0.01 | 99.93±0.01 | 3.87±0.02 | 34.75±0.01*** | 1.0±0.02 |
表6 菌株HBUT-D7和HBUT-D玉米浆培养基发酵数据
Table 6 Data on fermentation of strain HBUT-D7 and HBUT-D in corn syrup medium
| 菌株 Strain | D-乳酸糖酸转化率D-lactate yield/% | D-乳酸光纯D-lactate optical purty/% | D-乳酸生产强度D-lactate productivity/(g·L-1·h-1) | L-乳酸消耗速率Rate of L-lac consumption/(mg·L-1·h-1) | 初始L-乳酸浓度Initial L-lac concentration/(g·L-1) |
|---|---|---|---|---|---|
| HBUT-D | 97.64±0.01 | 99.22±0.02 | 4.24±0.04 | 10.41±0.01*** | 1.0±0.02 |
| HBUT-D7 | 96.89±0.01 | 99.93±0.01 | 3.87±0.02 | 34.75±0.01*** | 1.0±0.02 |
| 菌株 Strain | D-乳酸糖酸转化率D-lactate yield/% | D-乳酸光纯 D-lactate optical purty/% | L-乳酸消耗速率Rate of L-lactate consumption/(mg·L-1·h-1) | D-乳酸生产强度D-lactate productivity/(g·L-1·h-1) |
|---|---|---|---|---|
| HBUT-D | 61.82±0.01 | 99.01±0.01 | 6.87±0.02*** | 1.93 |
| HBUT-D7 | 60.43±0.01 | 99.99±0.01 | 17.18±0.01*** | 1.88 |
表7 菌株HBUT-D和HBUT-D7糖蜜培养基发酵数据
Table 7 Data on the fermentation of strain HBUT-D and HBUT-D7 in molasses medium
| 菌株 Strain | D-乳酸糖酸转化率D-lactate yield/% | D-乳酸光纯 D-lactate optical purty/% | L-乳酸消耗速率Rate of L-lactate consumption/(mg·L-1·h-1) | D-乳酸生产强度D-lactate productivity/(g·L-1·h-1) |
|---|---|---|---|---|
| HBUT-D | 61.82±0.01 | 99.01±0.01 | 6.87±0.02*** | 1.93 |
| HBUT-D7 | 60.43±0.01 | 99.99±0.01 | 17.18±0.01*** | 1.88 |
| [1] | 吕汉强, 赵文花, 李含婷, 等. 聚乳酸可降解地膜对绿洲灌区玉米产量和农田水热特性的影响[J]. 甘肃农业大学学报, 2022, 57(5): 72-79, 88. |
| Lyu HQ, Zhao WH, Li HT, et al. Effects of polylactic acid degradable film on maize yield and soil hydrothermal characteristics in oasis irrigation area[J]. J Gansu Agric Univ, 2022, 57(5): 72-79, 88. | |
| [2] |
王健群, 张斌. 聚乳酸-羟基乙酸共聚物微球在骨组织工程中的应用[J]. 口腔医学研究, 2020, 36(9): 817-820.
doi: 10.13701/j.cnki.kqyxyj.2020.09.005 |
| Wang JQ, Zhang B. Application of poly(lactic-co-glycolic acid)copolymer microspheres in bone tissue engineering[J]. J Oral Sci Res, 2020, 36(9): 817-820. | |
| [3] | 吴一帆. D-乳酸嵌段共聚物的合成设计及其对左旋聚乳酸结晶行为和性能的影响[D]. 成都: 西南交通大学, 2020. |
| Wu YF. Synthesis design of D-lactic acid block copolymer and investigation on crystallization behaviors and properties of block copolymer/poly(L-lactic acid)blends[D]. Chengdu: Southwest Jiaotong University, 2020. | |
| [4] |
Grabar TB, Zhou S, Shanmugam KT, et al. Methylglyoxal bypass identified as source of chiral contamination in l(+)and d(-)-lactate fermentations by recombinant Escherichia coli[J]. Biotechnol Lett, 2006, 28(19): 1527-1535.
doi: 10.1007/s10529-006-9122-7 pmid: 16868860 |
| [5] | 张媛, 王小艳, 陈博, 等. 基因工程改造植物乳杆菌生产光学纯D-乳酸[J]. 当代化工, 2019, 48(4): 674-678, 682. |
| Zhang Y, Wang XY, Chen B, et al. Genetically engineered Lactobacillus plantarum for production of optical pure D-lactic acid[J]. Contemp Chem Ind, 2019, 48(4): 674-678, 682. | |
| [6] | 李义, 周卫强, 刘海军, 等. 玉米浆及其制备方法和应用以及高光学纯度乳酸的生产方法: CN114181980A[P]. 2022-03-15. |
| Li Y, Zhou W, Liu H, et al. Corn steep liquor, preparation method and application thereof, and production method of high-optical-purity lactic acid: CN114181980A[P]. 2022-03-15. | |
| [7] | Alexandri M, Hübner D, Schneider R, et al. Towards efficient production of highly optically pure d-lactic acid from lignocellulosic hydrolysates using newly isolated lactic acid bacteria[J]. N Biotechnol, 2022, 72: 1-10. |
| [8] | Zhou SD, Iverson AG, Grayburn WS. Doubling the catabolic reducing power(NADH)output of Escherichia coli fermentation for production of reduced products[J]. Biotechnol Prog, 2010, 26(1): 45-51. |
| [9] | Nasr R, Akbari Eidgahi MR. Construction of a synthetically engineered nirB promoter for expression of recombinant protein in Escherichia coli[J]. Jundishapur J Microbiol, 2014, 7(7): e15942. |
| [10] | 田甜. 高效利用蔗糖产D-乳酸工程菌的构建及其发酵研究[D]. 武汉: 湖北工业大学, 2013. |
| Tian T. Construction and fermentation of engineering bacteria producing D- lactic acid from sucrose with high efficiency[D]. Wuhan: Hubei University of Technology, 2013. | |
| [11] |
Kimura H, Futai M. Effects of phospholipids on L-lactate dehydrogenase from membranes of Escherichia coli. Activation and stabilization of the enzyme with phospholipids[J]. J Biol Chem, 1978, 253(4): 1095-1110.
pmid: 342518 |
| [12] |
刘红雨, 刘友存, 孟丽红, 等. 熵权法在水资源与水环境评价中的研究进展[J]. 冰川冻土, 2022, 44(1): 299-306.
doi: 10.7522/j.issn.1000-0240.2021.0131 |
|
Liu HY, Liu YC, Meng LH, et al. Research progress of entropy weight method in water resources and water environment[J]. J Glaciol Geocryol, 2022, 44(1): 299-306.
doi: 10.7522/j.issn.1000-0240.2021.0131 |
|
| [13] |
Aguilera L, Campos E, Giménez R, et al. Dual role of LldR in regulation of the lldPRD operon, involved in L-lactate metabolism in Escherichia coli[J]. J Bacteriol, 2008, 190(8): 2997-3005.
doi: 10.1128/JB.02013-07 pmid: 18263722 |
| [14] |
Futai M, Kimura H. Inducible membrane-bound L-lactate dehydrogenase from Escherichia coli. Purification and properties[J]. J Biol Chem, 1977, 252(16): 5820-5827.
pmid: 18473 |
| [15] | Bekker M, Alexeeva S, Laan W, et al. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool[J]. J Bacteriol, 2010, 192(3): 746-754. |
| [16] | 许琼丹. 基于糖转运及代谢相关基因敲除的产D-乳酸大肠杆菌工程菌的构建[D]. 武汉: 湖北工业大学, 2020. |
| Xu QD. Construction of D-lactic acid-producing Escherichia coli strains by knockout of sugar transport and metabolism genes[D]. Wuhan: Hubei University of Technology, 2020. | |
| [17] | 鲁春阳, 文枫, 杨庆媛, 等. 基于改进TOPSIS法的城市土地利用绩效评价及障碍因子诊断——以重庆市为例[J]. 资源科学, 2011, 33(3): 535-541. |
| Lu CY, Wen F, Yang QY, et al. An evaluation of urban land use performance based on the improved TOPSIS method and diagnosis of its obstacle indicators: a case study of Chongqing[J]. Resour Sci, 2011, 33(3): 535-541. | |
| [18] | 张帆, 陈梦茹, 邢英英, 等. 基于熵权法和TOPSIS对马铃薯施肥和滴灌量组合的优化[J]. 植物营养与肥料学报, 2023, 29(4): 732-744. |
| Zhang F, Chen MR, Xing YY, et al. Optimization of fertilizer and drip irrigation levels for efficient potato production based on entropy weight method and TOPSIS[J]. J Plant Nutr Fertil, 2023, 29(4): 732-744. | |
| [19] | Fantino JR, Py B, Fontecave M, et al. A genetic analysis of the response of Escherichia coli to cobalt stress[J]. Environ Microbiol, 2010, 12(10): 2846-2857. |
| [20] |
Zaslaver A, Mayo AE, Rosenberg R, et al. Just-in-time transcription program in metabolic pathways[J]. Nat Genet, 2004, 36(5): 486-491.
pmid: 15107854 |
| [21] |
张玲, 林荣, 宋祖坤, 等. 启动子串联及改造提高FAD为辅基的葡萄糖脱氢酶在Bacillus subtilis中的表达[J]. 食品与发酵工业, 2019, 45(8): 15-21.
doi: 10.13995/j.cnki.11-1802/ts.018685 |
| Zhang L, Lin R, Song ZK, et al. Promoter tandem and transformation for FAD-conjugated glucose dehydrogenase expression in Bacillus subtilis[J]. Food Ferment Ind, 2019, 45(8): 15-21. | |
| [22] | 葛春蕾, 刘中美, 崔文璟, 等. 通过串联启动子实现纳豆激酶在枯草芽孢杆菌中的高效表达[J]. 现代食品科技, 2016, 32(11): 72-77, 15. |
| Ge CL, Liu ZM, Cui WJ, et al. Efficient overexpression of recombinant nattokinase in Bacillus subtilis by tandem promoters[J]. Mod Food Sci Technol, 2016, 32(11): 72-77, 15. | |
| [23] | Chauhan V, Bahrudeen MNM, Palma CSD, et al. Analytical kinetic model of native tandem promoters in E. coli[J]. PLoS Comput Biol, 2022, 18(1): e1009824. |
| [24] | Kasho K, Ozaki S, Katayama T. IHF and fis as Escherichia coli cell cycle regulators: activation of the replication origin oriC and the regulatory cycle of the DnaA initiator[J]. Int J Mol Sci, 2023, 24(14): 11572. |
| [25] |
Kim D, Seo SW, Gao Y, et al. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP[J]. Nucleic Acids Res, 2018, 46(6): 2901-2917.
doi: 10.1093/nar/gky069 pmid: 29394395 |
| [1] | 杨红艳, 韩筱, 杨建军. pDNA质粒在一次性生物反应器中的放大生产研究[J]. 生物技术通报, 2024, 40(1): 168-175. |
| [2] | 陈彩萍, 任昊, 龙腾飞, 何冰, 鲁兆祥, 孙坚. 大肠杆菌Nissle 1917对炎症性肠病治疗作用的研究进展[J]. 生物技术通报, 2023, 39(6): 109-118. |
| [3] | 吴莉丹, 冉雪琴, 牛熙, 黄世会, 李升, 王嘉福. 猪源致病性大肠杆菌基因组比较与毒力因子分析[J]. 生物技术通报, 2023, 39(12): 287-299. |
| [4] | 侯炜辰, 叶柯, 李洁, 张洋子, 许文涛, 朱龙佼, 李相阳. 基于抗体-适配体夹心生物传感器检测大肠杆菌O157: H7[J]. 生物技术通报, 2023, 39(12): 81-89. |
| [5] | 李奕雅, 吴一凡, 丁能水, 范小萍, 陈凡. 荧光素酶辅助定量大肠杆菌破碎效果的方法[J]. 生物技术通报, 2023, 39(12): 90-98. |
| [6] | 唐瑞琪, 赵心清, 朱笃, 汪涯. 大肠杆菌对木质纤维素水解液抑制物的胁迫耐受性[J]. 生物技术通报, 2023, 39(11): 205-216. |
| [7] | 李仁瀚, 张乐乐, 刘春立, 刘秀霞, 白仲虎, 杨艳坤, 李业. 基于紫色杆菌素生物合成途径的L-色氨酸生物传感器的构建[J]. 生物技术通报, 2023, 39(10): 80-92. |
| [8] | 高伟欣, 黄火清, 赵晶, 张鑫, 杨宁, 杨浩萌. 应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J]. 生物技术通报, 2022, 38(8): 60-68. |
| [9] | 孙曼銮, 葛赛, 卜佳, 朱壮彦. 大肠杆菌核糖核酸酶调控机制研究[J]. 生物技术通报, 2022, 38(3): 234-245. |
| [10] | 李晓芳, 刘慧燕, 潘琳, 艾治宇, 李一鸣, 张恒, 方海田. 常温常压等离子体诱变选育高产L-异亮氨酸大肠杆菌[J]. 生物技术通报, 2022, 38(1): 150-156. |
| [11] | 吴蓉, 曹佳睿, 曹君, 刘飞翔, 杨猛, 苏二正. 南极假丝酵母脂肪酶B基因在大肠杆菌中的表达和发酵优化[J]. 生物技术通报, 2021, 37(2): 138-148. |
| [12] | 王凯凯, 王晓璐, 苏小运, 张杰. 大肠杆菌双质粒CRISPR-Cas9系统的优化及应用[J]. 生物技术通报, 2021, 37(12): 252-264. |
| [13] | 陈桥, 吴海英, 王宗寿, 谢雨康, 李宜青, 孙俊松. 聚羟基丁酸酯合成引发的高密度生长大肠杆菌的多位点突变分析[J]. 生物技术通报, 2020, 36(7): 112-118. |
| [14] | 张春晨, 胡双艳, 阮海华. 人源溶菌酶在大肠杆菌中的表达与复性研究[J]. 生物技术通报, 2020, 36(3): 153-161. |
| [15] | 王琦, 颜春蕾, 高洪伟, 吴薇, 杨庆利. 基于核酸适配体传感器检测食品致病菌的研究进展[J]. 生物技术通报, 2020, 36(11): 245-258. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||