








生物技术通报 ›› 2023, Vol. 39 ›› Issue (11): 205-216.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0722
收稿日期:2023-07-28
出版日期:2023-11-26
发布日期:2023-12-20
通讯作者:
汪涯,男,博士,副教授,研究方向:微生物生态和代谢调控;E-mail: wangya@jxstnu.edu.cn作者简介:唐瑞琪,女,博士,讲师,研究方向:微生物代谢工程和合成生物学;E-mail: rq_tang@jxstnu.edu.cn
基金资助:
TANG Rui-qi1( ), ZHAO Xin-qing2, ZHU Du1, WANG Ya1(
), ZHAO Xin-qing2, ZHU Du1, WANG Ya1( )
)
Received:2023-07-28
Published:2023-11-26
Online:2023-12-20
摘要:
木质纤维素类生物质是前景广阔的化石原料替代品,其生物炼制可生产生物能源、生物基化学品和生物材料等多种产品,可降低碳排放,有助于实现“双碳”目标,因此受到越来越多的关注。然而,木质纤维素生物炼制需要经过预处理、微生物发酵和产物纯化等多个步骤,其中,预处理过程产生的多种化合物抑制微生物的细胞生长和发酵性能,是制约生物转化效率的瓶颈之一。大肠杆菌是木质纤维素生物炼制常用的宿主,被广泛应用于多种化合物的生产,研究其对木质纤维素水解液中抑制物的耐受性,对于提高木质纤维素生物炼制效率具有重要意义。本文首先介绍了木质纤维素的主要成分和基本结构,对木质纤维素的预处理方法以及预处理后水解液中的主要抑制物种类进行了简单阐述;随后,总结了木质纤维素水解液中几类主要抑制物呋喃类、羧酸类和酚类对大肠杆菌细胞的毒性,以及大肠杆菌对上述抑制物的胁迫响应机制和基于机制的菌株改造靶点;最后,综述了提高大肠杆菌对上述抑制物的胁迫耐受性的菌株改造策略,包括随机突变、实验室适应性进化和组学辅助的理性设计等,为利用代谢工程构建用于木质纤维素生物炼制的高效大肠杆菌菌株提供参考。
唐瑞琪, 赵心清, 朱笃, 汪涯. 大肠杆菌对木质纤维素水解液抑制物的胁迫耐受性[J]. 生物技术通报, 2023, 39(11): 205-216.
TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates[J]. Biotechnology Bulletin, 2023, 39(11): 205-216.

图1 木质纤维素水解液中常见的抑制物及其来源 HMF *表示5-羟甲基糠醛
Fig. 1 Common inhibitors in lignocellulosic hydrolysates and their sources HMF * stands for 5-hydroxylmethylfurfural
| 基因 Gene | 敲除/过表达 Deletion/Overexpression | 描述 Description | 抑制物 Inhibitor | 参考文献 Reference |
|---|---|---|---|---|
| pgi, encoding glucose-6-phosphate isomerase | Deletion | Shunt glucose to pentose phosphate pathway to increase NADPH production | Furfural, HMF | [ |
| pntAB, encoding pyridine nucleotide transhydrogenase | Overexpression | Convert NADP+ to NADPH using PntAB to increase NADPH regeneration | Furfural, HMF | [ |
| yqhD, encoding aldehyde reductase | Deletion | Delete NADPH-dependent YqhD to reduce NADPH consumption | Furfural, HMF | [ |
| yqhC, encoding transcriptional activator | Deletion | Delete YqhC to downregulate yqhDexpression, reducing NADPH consumption | Furfural | [ |
| fucO, encoding propanediol oxidoreductase | Overexpression | Convert furfural using NADH-dependent FucO to reduce NADPH consumption | Furfural | [ |
| yghA, encoding oxidoreductase | Overexpression | Convert furfural using NADH-dependent YghA to reduce NADPH consumption | Furfural, HMF | [ |
| pncB and nadE, encoding NAD salvage pathway enzymes | Overexpression | Increase NAD(P)H level through the nicotine amide salvage pathway | Furfural | [ |
| Heterologous xylBand BaBAD, encoding benzyl alcohol dehydrogenases | Overexpression | Convert furfural using NADH-dependent XylB and BaBAD to reduce NADPH consumption | Furfural | [ |
| thyA, encoding thymidylate synthase | Overexpression | Overexpress ThyA to increase dTMP level for DNA repair | Furfural | [ |
| potE and puuP, encoding polyamine transporters | Overexpression | Increase cytoplasmic polyamine level to maintain DNA synthesis | Furfural | [ |
| lpcA, encoding D-sedoheptulose-7-phosphate isomerase | Overexpression | Overexpress LpcA to increase formation of lipopolysaccharides and NADPH | Furfural | [ |
| pssA, encoding phosphatidylserine synthase | Overexpression | Increase phosphatidylethanolamine content to increase membrane integrity | Furfural, HMF | [ |
| mdtJI, encoding multidrug resistance efflux pump | Overexpression | Export furfural by efflux pump MdtJI | Furfural, HMF | [ |
| groESL, encoding chaperonin | Overexpression | Maintain proper folding of proteins | Furfural | [ |
表1 提高大肠杆菌呋喃类抑制物耐受性的基因靶点
Table 1 Gene targets for improving the tolerance of E. coli to furan inhibitors
| 基因 Gene | 敲除/过表达 Deletion/Overexpression | 描述 Description | 抑制物 Inhibitor | 参考文献 Reference |
|---|---|---|---|---|
| pgi, encoding glucose-6-phosphate isomerase | Deletion | Shunt glucose to pentose phosphate pathway to increase NADPH production | Furfural, HMF | [ |
| pntAB, encoding pyridine nucleotide transhydrogenase | Overexpression | Convert NADP+ to NADPH using PntAB to increase NADPH regeneration | Furfural, HMF | [ |
| yqhD, encoding aldehyde reductase | Deletion | Delete NADPH-dependent YqhD to reduce NADPH consumption | Furfural, HMF | [ |
| yqhC, encoding transcriptional activator | Deletion | Delete YqhC to downregulate yqhDexpression, reducing NADPH consumption | Furfural | [ |
| fucO, encoding propanediol oxidoreductase | Overexpression | Convert furfural using NADH-dependent FucO to reduce NADPH consumption | Furfural | [ |
| yghA, encoding oxidoreductase | Overexpression | Convert furfural using NADH-dependent YghA to reduce NADPH consumption | Furfural, HMF | [ |
| pncB and nadE, encoding NAD salvage pathway enzymes | Overexpression | Increase NAD(P)H level through the nicotine amide salvage pathway | Furfural | [ |
| Heterologous xylBand BaBAD, encoding benzyl alcohol dehydrogenases | Overexpression | Convert furfural using NADH-dependent XylB and BaBAD to reduce NADPH consumption | Furfural | [ |
| thyA, encoding thymidylate synthase | Overexpression | Overexpress ThyA to increase dTMP level for DNA repair | Furfural | [ |
| potE and puuP, encoding polyamine transporters | Overexpression | Increase cytoplasmic polyamine level to maintain DNA synthesis | Furfural | [ |
| lpcA, encoding D-sedoheptulose-7-phosphate isomerase | Overexpression | Overexpress LpcA to increase formation of lipopolysaccharides and NADPH | Furfural | [ |
| pssA, encoding phosphatidylserine synthase | Overexpression | Increase phosphatidylethanolamine content to increase membrane integrity | Furfural, HMF | [ |
| mdtJI, encoding multidrug resistance efflux pump | Overexpression | Export furfural by efflux pump MdtJI | Furfural, HMF | [ |
| groESL, encoding chaperonin | Overexpression | Maintain proper folding of proteins | Furfural | [ |
| 基因 Gene | 描述 Description | 胁迫 Stress | 参考文献 Reference |
|---|---|---|---|
| dsrA and hfq, encoding small noncoding RNA and chaperone | DsrA increases rpoS mRNA stability and activate RpoS translation, Hfq promotes DsrA annealing to the rpoS5' untranscribed region(UTR) | Low pH | [ |
| Heterologous cfaS, encoding cyclopropane fatty acid synthase | Decrease membrane permeability and fluidity | Low pH | [ |
| Heterologous cbpA, encoding chaperone | CbpA plays a role in protein and DNA repair | Acetate | [ |
| gadE, encoding transcriptional activator | GadE activates acid resistance system | Low pH | [ |
| hdeB, encoding periplasmic acid stress chaperone | HdeB prevents periplasmic proteins aggregation at low pH | Low pH | [ |
| sodB and katE, encoding superoxide dismutase and catalase | SodB and KatE are ROS scavengers | Low pH | [ |
表2 提高大肠杆菌耐酸性的过表达靶点
Table 2 Overexpression targets for improving the tolerance of E. coli to acid
| 基因 Gene | 描述 Description | 胁迫 Stress | 参考文献 Reference |
|---|---|---|---|
| dsrA and hfq, encoding small noncoding RNA and chaperone | DsrA increases rpoS mRNA stability and activate RpoS translation, Hfq promotes DsrA annealing to the rpoS5' untranscribed region(UTR) | Low pH | [ |
| Heterologous cfaS, encoding cyclopropane fatty acid synthase | Decrease membrane permeability and fluidity | Low pH | [ |
| Heterologous cbpA, encoding chaperone | CbpA plays a role in protein and DNA repair | Acetate | [ |
| gadE, encoding transcriptional activator | GadE activates acid resistance system | Low pH | [ |
| hdeB, encoding periplasmic acid stress chaperone | HdeB prevents periplasmic proteins aggregation at low pH | Low pH | [ |
| sodB and katE, encoding superoxide dismutase and catalase | SodB and KatE are ROS scavengers | Low pH | [ |
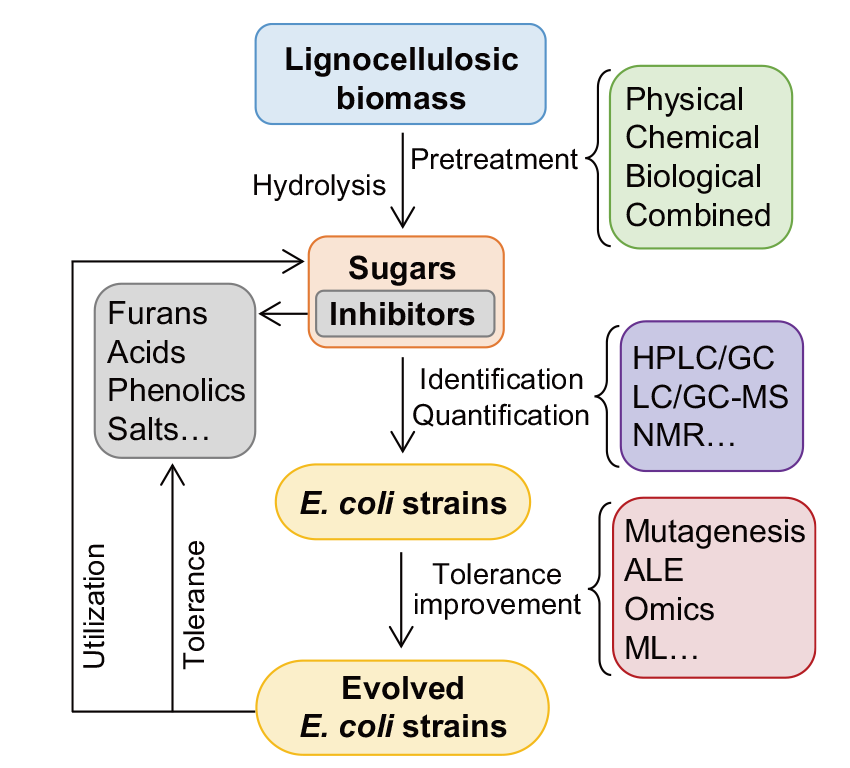
图3 提高大肠杆菌对木质纤维素水解液中抑制物耐受性的示意图 HPLC/GC表示高效液相色谱或气相色谱,LC-/GC-MS表示液相或气相色谱与质谱联用,NMR表示核磁共振,ALE和ML分别表示实验室适应性进化和机器学习
Fig. 3 Schematic diagram of the tolerance improvement of E. coli to inhibitors in lignocellulosic hydrolysates HPLC/GC indicate high-performance liquid chromatography/gas chromatography, LC-/GC-MS indicate liquid chromatography-/gas chromatography-mass spectrometer, NMR indicates nuclear magnetic resonance, ALE indicates adaptive laboratory evolution, ML indicates machine learning
| [1] |
Lu HD, Yadav V, Bilal M, et al. Bioprospecting microbial hosts to valorize lignocellulose biomass - Environmental perspectives and value-added bioproducts[J]. Chemosphere, 2022, 288(Pt 2): 132574.
doi: 10.1016/j.chemosphere.2021.132574 URL |
| [2] |
Reshmy R, Philip E, Madhavan A, et al. Lignocellulose in future biorefineries: strategies for cost-effective production of biomaterials and bioenergy[J]. Bioresour Technol, 2022, 344(Pt B): 126241.
doi: 10.1016/j.biortech.2021.126241 URL |
| [3] |
Zhai R, Hu JG, Jin MJ. Towards efficient enzymatic saccharification of pretreated lignocellulose: enzyme inhibition by lignin-derived phenolics and recent trends in mitigation strategies[J]. Biotechnol Adv, 2022, 61: 108044.
doi: 10.1016/j.biotechadv.2022.108044 URL |
| [4] |
Tan JY, Li Y, Tan X, et al. Advances in pretreatment of straw biomass for sugar production[J]. Front Chem, 2021, 9: 696030.
doi: 10.3389/fchem.2021.696030 URL |
| [5] |
Zhao CH, Zhang YP, Li Y. Production of fuels and chemicals from renewable resources using engineered Escherichia coli[J]. Biotechnol Adv, 2019, 37(7): 107402.
doi: 10.1016/j.biotechadv.2019.06.001 URL |
| [6] | Banerjee S, Pandit C, Gundupalli MP, et al. Life cycle assessment of revalorization of lignocellulose for the development of biorefineries[J]. Environ Dev Sustain, 2023. https://doi.org/10.1007/s10668-023-03360-4. |
| [7] |
Zhou M, Tian XJ. Development of different pretreatments and related technologies for efficient biomass conversion of lignocellulose[J]. Int J Biol Macromol, 2022, 202: 256-268.
doi: 10.1016/j.ijbiomac.2022.01.036 pmid: 35032493 |
| [8] |
Haldar D, Purkait MK. A review on the environment-friendly emerging techniques for pretreatment of lignocellulosic biomass: Mechanistic insight and advancements[J]. Chemosphere, 2021, 264(Pt 2): 128523.
doi: 10.1016/j.chemosphere.2020.128523 URL |
| [9] |
Shen XJ, Sun RC. Recent advances in lignocellulose prior-fractionation for biomaterials, biochemicals, and bioenergy[J]. Carbohydr Polym, 2021, 261: 117884.
doi: 10.1016/j.carbpol.2021.117884 URL |
| [10] |
Zhang R, Gao HR, Wang YT, et al. Challenges and perspectives of green-like lignocellulose pretreatments selectable for low-cost biofuels and high-value bioproduction[J]. Bioresour Technol, 2023, 369: 128315.
doi: 10.1016/j.biortech.2022.128315 URL |
| [11] |
van der Pol EC, Bakker RR, Baets P, et al. By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for(bio)chemicals and fuels[J]. Appl Microbiol Biotechnol, 2014, 98(23): 9579-9593.
doi: 10.1007/s00253-014-6158-9 URL |
| [12] |
Gallego-García M, Moreno AD, Manzanares P, et al. Recent advances on physical technologies for the pretreatment of food waste and lignocellulosic residues[J]. Bioresour Technol, 2023, 369: 128397.
doi: 10.1016/j.biortech.2022.128397 URL |
| [13] |
Shukla A, Kumar D, Girdhar M, et al. Strategies of pretreatment of feedstocks for optimized bioethanol production: distinct and integrated approaches[J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 44.
doi: 10.1186/s13068-023-02295-2 |
| [14] | Wu ZY, Peng K, Zhang Y, et al. Lignocellulose dissociation with biological pretreatment towards the biochemical platform: a review[J]. Mater Today Bio, 2022, 16: 100445. |
| [15] |
Sai Bharadwaj AVSL, Dev S, Zhuang JS, et al. Review of chemical pretreatment of lignocellulosic biomass using low-liquid and low-chemical catalysts for effective bioconversion[J]. Bioresour Technol, 2023, 368: 128339.
doi: 10.1016/j.biortech.2022.128339 URL |
| [16] |
Shan WW, Yan YL, Li YD, et al. Microbial tolerance engineering for boosting lactic acid production from lignocellulose[J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 78.
doi: 10.1186/s13068-023-02334-y |
| [17] |
Balasundaram G, Banu R, Varjani S, et al. Recalcitrant compounds formation, their toxicity, and mitigation: key issues in biomass pretreatment and anaerobic digestion[J]. Chemosphere, 2022, 291(Pt 3): 132930.
doi: 10.1016/j.chemosphere.2021.132930 URL |
| [18] |
Guo HL, Zhao Y, Chang JS, et al. Inhibitor formation and detoxification during lignocellulose biorefinery: a review[J]. Bioresour Technol, 2022, 361: 127666.
doi: 10.1016/j.biortech.2022.127666 URL |
| [19] |
Ujor VC, Okonkwo CC. Microbial detoxification of lignocellulosic biomass hydrolysates: biochemical and molecular aspects, challenges, exploits and future perspectives[J]. Front Bioeng Biotechnol, 2022, 10: 1061667.
doi: 10.3389/fbioe.2022.1061667 URL |
| [20] |
Gutiérrez T, Ingram LO, Preston JF. Purification and characterization of a furfural reductase(FFR)from Escherichia coli strain LYO1- an enzyme important in the detoxification of furfural during ethanol production[J]. J Biotechnol, 2006, 121(2): 154-164.
pmid: 16111779 |
| [21] |
Miller EN, Jarboe LR, Yomano LP, et al. Silencing of NADPH-dependent oxidoreductase genes(yqhD and dkgA)in furfural-resistant ethanologenic Escherichia coli[J]. Appl Environ Microbiol, 2009, 75(13): 4315-4323.
doi: 10.1128/AEM.00567-09 URL |
| [22] |
Miller EN, Jarboe LR, Turner PC, et al. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180[J]. Appl Environ Microbiol, 2009, 75(19): 6132-6141.
doi: 10.1128/AEM.01187-09 URL |
| [23] | Hadi SM, Shahabuddin, Rehman A. Specificity of the interaction of furfural with DNA[J]. Mutat Res, 1989, 225(3): 101-106. |
| [24] |
Khan QA, Shamsi FA, Hadi SM. Mutagenicity of furfural in plasmid DNA[J]. Cancer Lett, 1995, 89(1): 95-99.
pmid: 7882307 |
| [25] |
Wang JQ, Zhang Y, Chen YL, et al. Global regulator engineering significantly improved Escherichia coli tolerances toward inhibitors of lignocellulosic hydrolysates[J]. Biotechnol Bioeng, 2012, 109(12): 3133-3142.
doi: 10.1002/bit.v109.12 URL |
| [26] |
Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium[J]. Microbiol Rev, 1991, 55(4): 561-585.
doi: 10.1128/mr.55.4.561-585.1991 pmid: 1779927 |
| [27] |
Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium[J]. Nat Rev Microbiol, 2013, 11(7): 443-454.
doi: 10.1038/nrmicro3032 pmid: 23712352 |
| [28] |
Boopathy R, Bokang H, Daniels L. Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria[J]. J Ind Microbiol, 1993, 11(3): 147-150.
doi: 10.1007/BF01583715 URL |
| [29] |
Miller EN, Turner PC, Jarboe LR, et al. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180[J]. Biotechnol Lett, 2010, 32(5): 661-667.
doi: 10.1007/s10529-010-0209-9 pmid: 20131081 |
| [30] |
Shahabuddin, Rahman A, Hadi SM. Specificity of the in vitro interaction of methylfurfural with DNA[J]. Mutagenesis, 1990, 5(2): 131-136.
pmid: 2140425 |
| [31] |
Jilani SB, Dev C, Eqbal D, et al. Deletion of pgi gene in E. coli increases tolerance to furfural and 5-hydroxymethyl furfural in media containing glucose-xylose mixture[J]. Microb Cell Fact, 2020, 19(1): 153.
doi: 10.1186/s12934-020-01414-0 pmid: 32723338 |
| [32] |
Turner PC, Miller EN, Jarboe LR, et al. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance[J]. J Ind Microbiol Biotechnol, 2011, 38(3): 431-439.
doi: 10.1007/s10295-010-0787-5 URL |
| [33] |
Wang X, Miller EN, Yomano LP, et al. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate[J]. Appl Environ Microbiol, 2011, 77(15): 5132-5140.
doi: 10.1128/AEM.05008-11 URL |
| [34] |
Jilani SB, Prasad R, Yazdani SS. Overexpression of oxidoreductase YghA confers tolerance of furfural in ethanologenic Escherichia coli strain SSK42[J]. Appl Environ Microbiol, 2021, 87(23): e0185521.
doi: 10.1128/AEM.01855-21 URL |
| [35] |
Song HS, Jeon JM, Kim HJ, et al. Increase in furfural tolerance by combinatorial overexpression of NAD salvage pathway enzymes in engineered isobutanol-producing E. coli[J]. Bioresour Technol, 2017, 245(Pt B): 1430-1435.
doi: 10.1016/j.biortech.2017.05.197 URL |
| [36] |
Willson BJ, Herman R, Langer S, et al. Improved furfural tolerance in Escherichia coli mediated by heterologous NADH-dependent benzyl alcohol dehydrogenases[J]. Biochem J, 2022, 479(10): 1045-1058.
doi: 10.1042/BCJ20210811 URL |
| [37] |
Zheng HB, Wang X, Yomano LP, et al. Increase in furfural tolerance in ethanologenic Escherichia coli LY180 by plasmid-based expression of thyA[J]. Appl Environ Microbiol, 2012, 78(12): 4346-4352.
doi: 10.1128/AEM.00356-12 URL |
| [38] |
Geddes RD, Wang X, Yomano LP, et al. Polyamine transporters and polyamines increase furfural tolerance during xylose fermentation with ethanologenic Escherichia coli strain LY180[J]. Appl Environ Microbiol, 2014, 80(19): 5955-5964.
doi: 10.1128/AEM.01913-14 URL |
| [39] |
Glebes TY, Sandoval NR, Reeder PJ, et al. Genome-wide mapping of furfural tolerance genes in Escherichia coli[J]. PLoS One, 2014, 9(1): e87540.
doi: 10.1371/journal.pone.0087540 URL |
| [40] |
Tan ZG, Khakbaz P, Chen YX, et al. Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables[J]. Metab Eng, 2017, 44: 1-12.
doi: 10.1016/j.ymben.2017.08.006 URL |
| [41] | Kurgan G, Panyon LA, Rodriguez-Sanchez Y, et al. Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli[J]. Appl Environ Microbiol, 2019, 85(6): e02985-e02918. |
| [42] |
Geiger LE, Morris DR. Polyamine deficiency reduces the rate of DNA replication fork movement in Escherichia coli[J]. Nature, 1978, 272(5655): 730-732.
doi: 10.1038/272730a0 |
| [43] |
Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli[J]. Biotechnol Biofuels, 2009, 2: 26.
doi: 10.1186/1754-6834-2-26 |
| [44] |
Sá-Pessoa J, Paiva S, Ribas D, et al. SATP(YaaH), a succinate-acetate transporter protein in Escherichia coli[J]. Biochem J, 2013, 454(3): 585-595.
doi: 10.1042/BJ20130412 pmid: 23844911 |
| [45] |
Gimenez R, Nuñez MF, Badia J, et al. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli[J]. J Bacteriol, 2003, 185(21): 6448-6455.
doi: 10.1128/JB.185.21.6448-6455.2003 pmid: 14563880 |
| [46] |
Roe AJ, McLaggan D, Davidson I, et al. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids[J]. J Bacteriol, 1998, 180(4): 767-772.
doi: 10.1128/JB.180.4.767-772.1998 pmid: 9473028 |
| [47] |
Walter A, Gutknecht J. Monocarboxylic acid permeation through lipid bilayer membranes[J]. J Membr Biol, 1984, 77(3): 255-264.
doi: 10.1007/BF01870573 URL |
| [48] |
Guan NZ, Liu L. Microbial response to acid stress: mechanisms and applications[J]. Appl Microbiol Biotechnol, 2020, 104(1): 51-65.
doi: 10.1007/s00253-019-10226-1 pmid: 31773206 |
| [49] | Sun YR. F1F0-ATPase functions under markedly acidic conditions in bacteria[M]// Regulation of Ca2+-ATPases, V-ATPases and F-ATPases. Cham: Springer, 2016: 459-468. |
| [50] |
Maurer LM, Yohannes E, Bondurant SS, et al. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12[J]. J Bacteriol, 2005, 187(1): 304-319.
doi: 10.1128/JB.187.1.304-319.2005 pmid: 15601715 |
| [51] |
Choi SH, Baumler DJ, Kaspar CW. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157: H7[J]. Appl Environ Microbiol, 2000, 66(9): 3911-3916.
doi: 10.1128/AEM.66.9.3911-3916.2000 URL |
| [52] |
Cherrington CA, Hinton M, Chopra I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli[J]. J Appl Bacteriol, 1990, 68(1): 69-74.
doi: 10.1111/jam.1990.68.issue-1 URL |
| [53] |
Roe AJ, O'Byrne C, McLaggan D, et al. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity[J]. Microbiology, 2002, 148(Pt 7): 2215-2222.
doi: 10.1099/00221287-148-7-2215 URL |
| [54] |
Brown JL, Ross T, McMeekin TA, et al. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance[J]. Int J Food Microbiol, 1997, 37(2/3): 163-173.
doi: 10.1016/S0168-1605(97)00068-8 URL |
| [55] |
Shabala L, Ross T. Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+[J]. Res Microbiol, 2008, 159(6): 458-461.
doi: 10.1016/j.resmic.2008.04.011 pmid: 18562182 |
| [56] | 李书廷, 洪坤强, 汪保卫, 等. 大肠杆菌乙酸耐受性菌株的构建及其耐受机制研究进展[J]. 微生物学通报, 2020, 47(12): 4250-4259. |
| Li ST, Hong KQ, Wang BW, et al. Advances in construction of acetic acid tolerance Escherichia coli[J]. Microbiol China, 2020, 47(12): 4250-4259. | |
| [57] | 郝雪雁, 刘梦晓, 韩紫依, 等. 大肠杆菌的耐酸机制及其改造研究进展[J]. 微生物学通报, 2023, 50(10):4667-4680. |
| Hao XY, Liu MX, Han ZY, et al. Advances in acid-resistant mechanisms and modifications of Escherichia coli[J]. Microbiol China, 2023, 50(10):4667-4680. | |
| [58] |
Mallick S, Das S. Acid-tolerant bacteria and prospects in industrial and environmental applications[J]. Appl Microbiol Biotechnol, 2023, 107(11): 3355-3374.
doi: 10.1007/s00253-023-12529-w |
| [59] |
Yang JH, Zhang J, Zhu ZM, et al. The challenges and prospects of Escherichia coli as an organic acid production host under acid stress[J]. Appl Microbiol Biotechnol, 2021, 105(21/22): 8091-8107.
doi: 10.1007/s00253-021-11577-4 |
| [60] |
Xu Y, Zhao Z, Tong WH, et al. An acid-tolerance response system protecting exponentially growing Escherichia coli[J]. Nat Commun, 2020, 11(1): 1496.
doi: 10.1038/s41467-020-15350-5 |
| [61] |
Kirkpatrick C, Maurer LM, Oyelakin NE, et al. Acetate and formate stress: opposite responses in the proteome of Escherichia coli[J]. J Bacteriol, 2001, 183(21): 6466-6477.
pmid: 11591692 |
| [62] | Kammel M, Pinske C, Sawers RG. FocA and its central role in fine-tuning pH homeostasis of enterobacterial formate metabolism[J]. Microbiology, 2022, 168(10). DOI: 10.1099/mic.0.001253. |
| [63] | Lin ZL, Li JH, Yan XF, et al. Engineering of the small noncoding RNA(sRNA)DsrA together with the sRNA chaperone Hfq enhances the acid tolerance of Escherichia coli[J]. Appl Environ Microbiol, 2021, 87(10): e02923-e02920. |
| [64] |
Hu WB, Tong YJ, Liu JJ, et al. Improving acid resistance of Escherichia coli base on the CfaS-mediated membrane engineering strategy derived from extreme acidophile[J]. Front Bioeng Biotechnol, 2023, 11: 1158931.
doi: 10.3389/fbioe.2023.1158931 URL |
| [65] |
Jiang ZM, Lu J, Tong YJ, et al. Enhancement of acid tolerance of Escherichia coli by introduction of molecule chaperone CbpA from extremophile[J]. World J Microbiol Biotechnol, 2023, 39(6): 158.
doi: 10.1007/s11274-023-03613-4 |
| [66] |
Yao XR, Liu P, Chen B, et al. Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH[J]. Microb Cell Fact, 2022, 21(1): 68.
doi: 10.1186/s12934-022-01795-4 |
| [67] |
Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli[J]. Biotechnol Bioeng, 1999, 65(1): 24-33.
doi: 10.1002/(sici)1097-0290(19991005)65:1<24::aid-bit4>3.0.co;2-2 pmid: 10440668 |
| [68] |
Fitzgerald DJ, Stratford M, Gasson MJ, et al. Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua[J]. J Appl Microbiol, 2004, 97(1): 104-113.
doi: 10.1111/j.1365-2672.2004.02275.x pmid: 15186447 |
| [69] | Pattrick CA, Webb JP, Green J, et al. Proteomic profiling, transcription factor modeling, and genomics of evolved tolerant strains elucidate mechanisms of vanillin toxicity in Escherichia coli[J]. mSystems, 2019, 4(4): e00163-e00119. |
| [70] |
Yu QH, Li YC, Wu B, et al. Novel mutagenesis and screening technologies for food microorganisms: advances and prospects[J]. Appl Microbiol Biotechnol, 2020, 104(4): 1517-1531.
doi: 10.1007/s00253-019-10341-z pmid: 31919586 |
| [71] |
Wu SR, Tian PF, Tan TW. Genomic landscapes of bacterial transposons and their applications in strain improvement[J]. Appl Microbiol Biotechnol, 2022, 106(19/20): 6383-6396.
doi: 10.1007/s00253-022-12170-z |
| [72] |
Zhang X, Zhang XF, Li HP, et al. Atmospheric and room temperature plasma(ARTP)as a new powerful mutagenesis tool[J]. Appl Microbiol Biotechnol, 2014, 98(12): 5387-5396.
doi: 10.1007/s00253-014-5755-y pmid: 24769904 |
| [73] |
Chen L, Xin QH, Ma LM, et al. Applications and research advance of genome shuffling for industrial microbial strains improvement[J]. World J Microbiol Biotechnol, 2020, 36(10): 158.
doi: 10.1007/s11274-020-02936-w |
| [74] | Yoon SH, Lee EG, Das A, et al. Enhanced vanillin production from recombinant E. coli using NTG mutagenesis and adsorbent resin[J]. Biotechnol Prog, 2007, 23(5): 1143-1148. |
| [75] |
Gao XX, Yang XF, Li JH, et al. Engineered global regulator H-NS improves the acid tolerance of E. coli[J]. Microb Cell Fact, 2018, 17(1): 118.
doi: 10.1186/s12934-018-0966-z |
| [76] |
Wang GL, Li Q, Zhang Z, et al. Recent progress in adaptive laboratory evolution of industrial microorganisms[J]. J Ind Microbiol Biotechnol, 2023, 50(1): kuac023.
doi: 10.1093/jimb/kuac023 URL |
| [77] |
Sandberg TE, Salazar MJ, Weng LL, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology[J]. Metab Eng, 2019, 56: 1-16.
doi: S1096-7176(19)30153-3 pmid: 31401242 |
| [78] |
Lu Q, Zhou XL, Liu JZ. Adaptive laboratory evolution and shuffling of Escherichia coli to enhance its tolerance and production of astaxanthin[J]. Biotechnol Biofuels Bioprod, 2022, 15(1): 17.
doi: 10.1186/s13068-022-02118-w |
| [79] |
Seong W, Han GH, Lim HS, et al. Adaptive laboratory evolution of Escherichia coli lacking cellular byproduct formation for enhanced acetate utilization through compensatory ATP consumption[J]. Metab Eng, 2020, 62: 249-259.
doi: 10.1016/j.ymben.2020.09.005 URL |
| [80] |
Warner JR, Reeder PJ, Karimpour-Fard A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nat Biotechnol, 2010, 28(8): 856-862.
doi: 10.1038/nbt.1653 pmid: 20639866 |
| [81] |
Glebes TY, Sandoval NR, Gillis JH, et al. Comparison of genome-wide selection strategies to identify furfural tolerance genes in Escherichia coli[J]. Biotechnol Bioeng, 2015, 112(1): 129-140.
doi: 10.1002/bit.v112.1 URL |
| [82] | Chang DD, Islam ZU, Zheng JF, et al. Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: the proteomics-guided metabolic engineering[J]. Synth Syst Biotechnol, 2021, 6(4): 384-395. |
| [83] |
Forsberg KJ, Patel S, Witt E, et al. Identification of genes conferring tolerance to lignocellulose-derived inhibitors by functional selections in soil metagenomes[J]. Appl Environ Microbiol, 2015, 82(2): 528-537.
doi: 10.1128/AEM.02838-15 URL |
| [84] |
Gurdo N, Volke DC, McCloskey D, et al. Automating the design-build-test-learn cycle towards next-generation bacterial cell factories[J]. N Biotechnol, 2023, 74: 1-15.
doi: 10.1016/j.nbt.2023.01.002 URL |
| [85] |
Phaneuf PV, Zielinski DC, Yurkovich JT, et al. Escherichia coli data-driven strain design using aggregated adaptive laboratory evolution mutational data[J]. ACS Synth Biol, 2021, 10(12): 3379-3395.
doi: 10.1021/acssynbio.1c00337 URL |
| [86] |
Choi TR, Song HS, Han YH, et al. Enhanced tolerance to inhibitors of Escherichia coli by heterologous expression of cyclopropane-fatty acid-acyl-phospholipid synthase(cfa)from Halomonas socia[J]. Bioprocess Biosyst Eng, 2020, 43(5): 909-918.
doi: 10.1007/s00449-020-02287-8 |
| [87] | Choi KR, Shin JH, Cho JS, et al. Systems metabolic engineering of Escherichia coli[J]. EcoSal Plus, 2016, 7(1). DOI: 10.1128/ecosalplus.ESP-0010-2015. |
| [88] |
Choi KR, Jang WD, Yang D, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering[J]. Trends Biotechnol, 2019, 37(8): 817-837.
doi: S0167-7799(19)30003-4 pmid: 30737009 |
| [89] |
Sandoval NR, Mills TY, Zhang M, et al. Elucidating acetate tolerance in E. coli using a genome-wide approach[J]. Metab Eng, 2011, 13(2): 214-224.
doi: 10.1016/j.ymben.2010.12.001 URL |
| [1] | 陈彩萍, 任昊, 龙腾飞, 何冰, 鲁兆祥, 孙坚. 大肠杆菌Nissle 1917对炎症性肠病治疗作用的研究进展[J]. 生物技术通报, 2023, 39(6): 109-118. |
| [2] | 吴莉丹, 冉雪琴, 牛熙, 黄世会, 李升, 王嘉福. 猪源致病性大肠杆菌基因组比较与毒力因子分析[J]. 生物技术通报, 2023, 39(12): 287-299. |
| [3] | 侯炜辰, 叶柯, 李洁, 张洋子, 许文涛, 朱龙佼, 李相阳. 基于抗体-适配体夹心生物传感器检测大肠杆菌O157: H7[J]. 生物技术通报, 2023, 39(12): 81-89. |
| [4] | 李奕雅, 吴一凡, 丁能水, 范小萍, 陈凡. 荧光素酶辅助定量大肠杆菌破碎效果的方法[J]. 生物技术通报, 2023, 39(12): 90-98. |
| [5] | 李昕悦, 周明海, 樊亚超, 廖莎, 张风丽, 刘晨光, 孙悦, 张霖, 赵心清. 基于转运蛋白工程提升微生物菌株耐受性和生物制造效率的研究进展[J]. 生物技术通报, 2023, 39(11): 123-136. |
| [6] | 胡锦超, 沈文琦, 徐超业, 樊雅祺, 卢浩宇, 蒋雯杰, 李世龙, 晋洪晨, 骆健美, 王敏. 微生物酸胁迫耐受性能强化的研究进展[J]. 生物技术通报, 2023, 39(11): 137-149. |
| [7] | 晏雄鹰, 王振, 王霞, 杨世辉. 微生物硫代谢与抗逆性[J]. 生物技术通报, 2023, 39(11): 150-167. |
| [8] | 王文韬, 冯颀, 刘晨光, 白凤武, 赵心清. 氧化还原敏感型基因元件增强酵母木质纤维素水解液抑制物胁迫耐受性[J]. 生物技术通报, 2023, 39(11): 360-372. |
| [9] | 李仁瀚, 张乐乐, 刘春立, 刘秀霞, 白仲虎, 杨艳坤, 李业. 基于紫色杆菌素生物合成途径的L-色氨酸生物传感器的构建[J]. 生物技术通报, 2023, 39(10): 80-92. |
| [10] | 高伟欣, 黄火清, 赵晶, 张鑫, 杨宁, 杨浩萌. 应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J]. 生物技术通报, 2022, 38(8): 60-68. |
| [11] | 孙曼銮, 葛赛, 卜佳, 朱壮彦. 大肠杆菌核糖核酸酶调控机制研究[J]. 生物技术通报, 2022, 38(3): 234-245. |
| [12] | 李晓芳, 刘慧燕, 潘琳, 艾治宇, 李一鸣, 张恒, 方海田. 常温常压等离子体诱变选育高产L-异亮氨酸大肠杆菌[J]. 生物技术通报, 2022, 38(1): 150-156. |
| [13] | 吴蓉, 曹佳睿, 曹君, 刘飞翔, 杨猛, 苏二正. 南极假丝酵母脂肪酶B基因在大肠杆菌中的表达和发酵优化[J]. 生物技术通报, 2021, 37(2): 138-148. |
| [14] | 王凯凯, 王晓璐, 苏小运, 张杰. 大肠杆菌双质粒CRISPR-Cas9系统的优化及应用[J]. 生物技术通报, 2021, 37(12): 252-264. |
| [15] | 顾翰琦, 邵玲智, 刘冉, 刘晓光, 李玲, 刘倩, 李洁, 张雅丽. 酿酒酵母酚类抑制物耐受性脂质组学研究[J]. 生物技术通报, 2021, 37(1): 15-23. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||