








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 191-202.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1161
收稿日期:2023-12-10
出版日期:2024-05-26
发布日期:2024-06-13
通讯作者:
周武,男,博士,副教授,研究方向:植物生理学;E-mail: zhouwu870624@163.com作者简介:李嘉欣,女,硕士研究生,研究方向:植物生态学;E-mail: 2544313591@qq.com
基金资助:
LI Jia-xin( ), LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu(
), LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu( )
)
Received:2023-12-10
Published:2024-05-26
Online:2024-06-13
摘要:
【目的】NRAMP(natural resistance-associated macrophage protein)基因家族是一类广泛存在于植物中的天然抗性相关巨噬蛋白,对植物中二价金属离子的细胞转运具有关键作用。为挖掘沙棘NRAMP基因家族响应重金属铅胁迫的关键基因,阐明沙棘响应重金属铅胁迫的分子机制。【方法】利用生物信息学技术鉴定沙棘NRAMP家族成员,对编码该家族蛋白的理化性质、系统进化、基因结构、保守基序、顺式作用元件、不同浓度铅胁迫下的NRAMP基因表达谱、种内种间共线性、蛋白互作网络进行综合分析。【结果】NRAMP基因家族的11个成员不均匀分布于7个染色体上,被分为Group1-Group3三大类群。NRAMP基因家族成员的启动子具有丰富的激素应答、分生组织表达和环境应激响应元件。转录组分析表明,沙棘NRAMP基因家族的11个成员在不同浓度的铅离子胁迫下呈现差异表达,其中8个基因在1 000 mg/kg的铅胁迫下显著下调,7个基因在高浓度铅胁迫(5 000mg/kg)下表达上调,并对选定的6个沙棘NRAMP基因进行了RT-qPCR验证。沙棘与单子叶植物小麦NRAMP基因家族的种间共线性比率高于双子叶植物拟南芥。蛋白质互作网络分析表明,沙棘NRAMP基因家族成员HrLNRAMP8可能主要通过乙烯途径调控重金属的吸收转运。【结论】在全基因组范围内从沙棘中系统鉴定得到11个HrLNRAMP家族成员。Group2亚族成员基因结构最复杂且一致性较低;所有的沙棘NRAMP成员都含有NRAMP家族特有的转运基序motif1。不同基因家族成员在铅胁迫条件下的表达具有偏好性,该基因家族通过响应重金属铅离子胁迫来调控基因表达水平,从而降低重金属胁迫带来的伤害。
李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202.
LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress[J]. Biotechnology Bulletin, 2024, 40(5): 191-202.
| 基因名称 Gene name | 基因ID Gene ID | 蛋白序列长度 Protein sequence length/aa | 分子量 Molecular weight/kD | 理论等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| HrLNRAMP1 | Hiprha1gene15634 | 512 | 56.21 | 4.89 | 质膜Plas |
| HrLNRAMP2 | Hiprha1gene30069 | 513 | 55.52 | 9.53 | 质膜Plas |
| HrLNRAMP3 | Hiprha1gene21177 | 626 | 69.13 | 8.50 | 质膜Plas |
| HrLNRAMP4 | Hiprha1gene09270 | 1 291 | 141.46 | 5.79 | 质膜Plas |
| HrLNRAMP5 | Hiprha1gene12503 | 530 | 58.39 | 4.93 | 质膜Plas |
| HrLNRAMP6 | Hiprha1gene10061 | 532 | 57.72 | 9.33 | 质膜Plas |
| HrLNRAMP7 | Hiprha1gene12853 | 538 | 58.68 | 5.94 | 质膜Plas |
| HrLNRAMP8 | Hiprha1gene11195 | 1 295 | 141.63 | 6.08 | 质膜Plas |
| HrLNRAMP9 | Hiprha1gene27351 | 513 | 56.33 | 4.73 | 质膜Plas |
| HrLNRAMP10 | Hiprha1gene30205 | 502 | 54.94 | 8.50 | 质膜Plas |
| HrLNRAMP11 | Hiprha1gene16210 | 327 | 36.24 | 6.40 | 液泡膜Vacu |
表1 沙棘NRAMP蛋白理化性质与亚细胞定位
Table 1 Physicochemical properties and subcellular localization of seabuckthorn NRAMP protein
| 基因名称 Gene name | 基因ID Gene ID | 蛋白序列长度 Protein sequence length/aa | 分子量 Molecular weight/kD | 理论等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| HrLNRAMP1 | Hiprha1gene15634 | 512 | 56.21 | 4.89 | 质膜Plas |
| HrLNRAMP2 | Hiprha1gene30069 | 513 | 55.52 | 9.53 | 质膜Plas |
| HrLNRAMP3 | Hiprha1gene21177 | 626 | 69.13 | 8.50 | 质膜Plas |
| HrLNRAMP4 | Hiprha1gene09270 | 1 291 | 141.46 | 5.79 | 质膜Plas |
| HrLNRAMP5 | Hiprha1gene12503 | 530 | 58.39 | 4.93 | 质膜Plas |
| HrLNRAMP6 | Hiprha1gene10061 | 532 | 57.72 | 9.33 | 质膜Plas |
| HrLNRAMP7 | Hiprha1gene12853 | 538 | 58.68 | 5.94 | 质膜Plas |
| HrLNRAMP8 | Hiprha1gene11195 | 1 295 | 141.63 | 6.08 | 质膜Plas |
| HrLNRAMP9 | Hiprha1gene27351 | 513 | 56.33 | 4.73 | 质膜Plas |
| HrLNRAMP10 | Hiprha1gene30205 | 502 | 54.94 | 8.50 | 质膜Plas |
| HrLNRAMP11 | Hiprha1gene16210 | 327 | 36.24 | 6.40 | 液泡膜Vacu |

图2 沙棘、拟南芥、水稻NRAMP蛋白的系统进化分析 不同分支颜色代表不同的亚族。红色三角标注的是沙棘NRAMP家族成员、蓝色五角星标注的是拟南芥NRAMP家族成员、黄色圆圈标注的是水稻NRAMP家族成员
Fig. 2 Phylogenetic analysis of NRAMP proteins in H. rhamnoides, A. thaliana and O. sativa Different branch colors indicate different subfamilies. Red triangles are labeled with H. rhamnoides NRAMP family members, blue pentagrams are labeled with Arabidopsis thaliana NRAMP family members, and yellow circles are labeled with Oryza sativa NRAMP family members

图3 沙棘NRAMP蛋白的多重比对 颜色越深,序列一致性程度越高
Fig. 3 Multiple alignment of H. rhamnoides NRAMP family proteins The darker the color, the more consistent the sequence
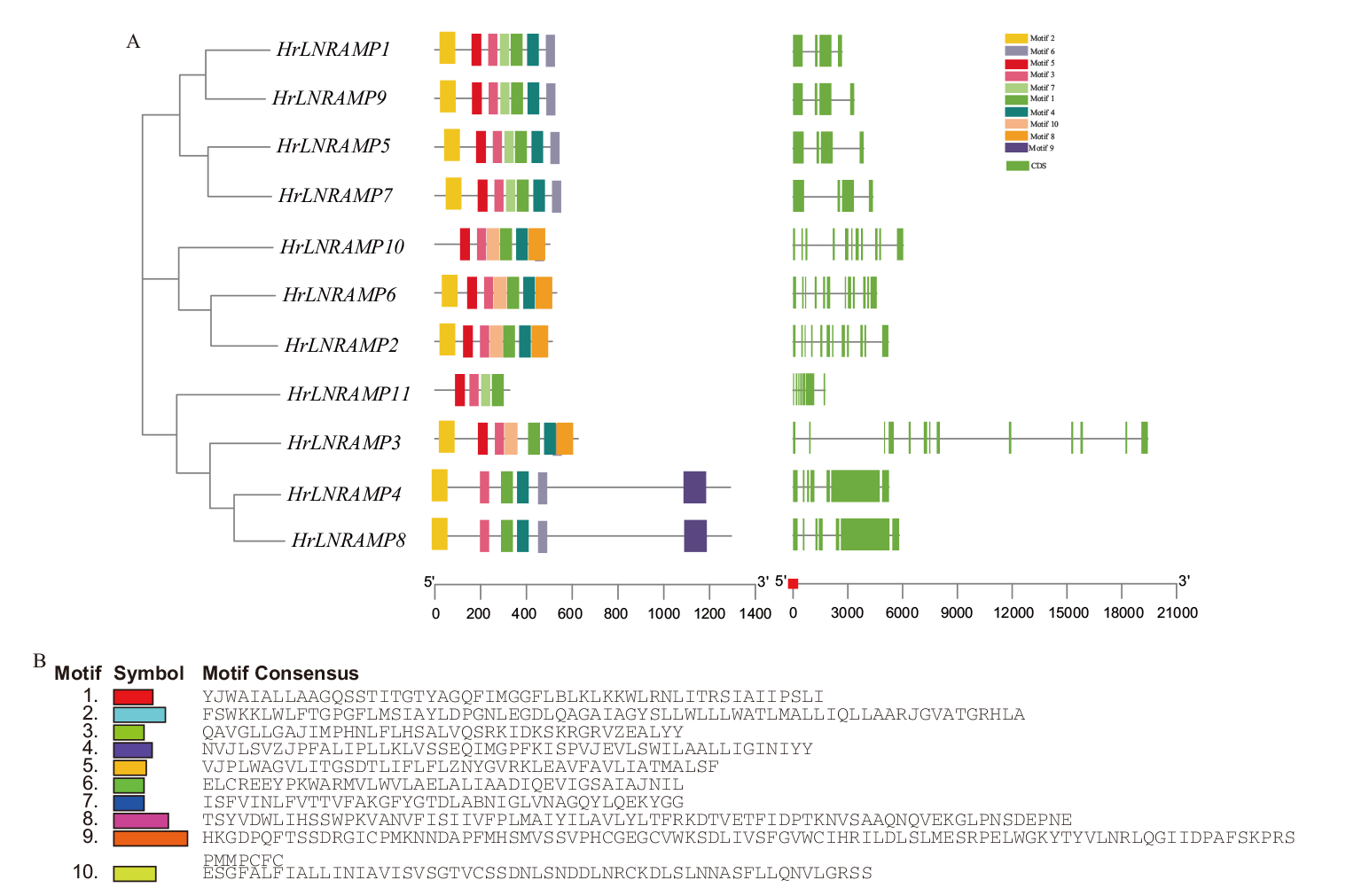
图4 NRAMP家族外显子-内含子与motif分析 A:沙棘NRAMP家族结构与保守基序;B:沙棘NRAMP家族保守基序组成
Fig. 4 NRAMP family exon-intron and motif analysis A: Structure and conserved motifs of the NRAMP family in seabuckthorn. B: Conserved motif composition of the NRAMP family in seabuckthorn
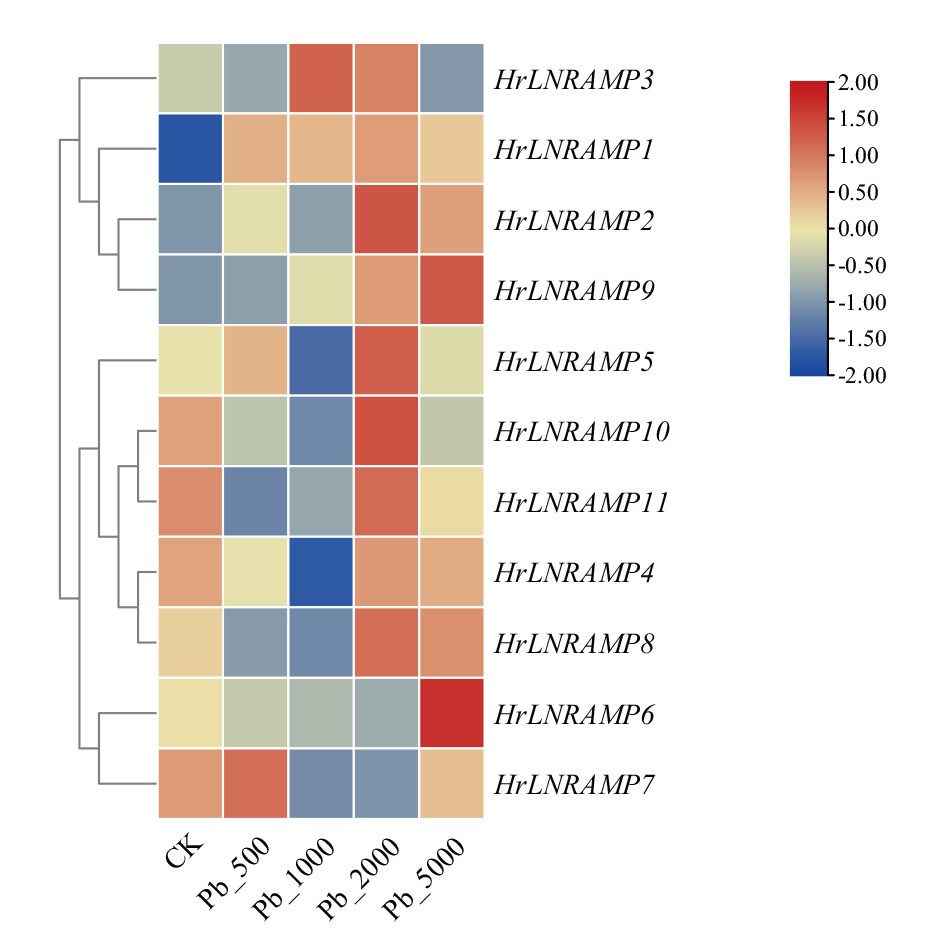
图6 沙棘NRAMP基因在不同浓度铅胁迫下的表达分析 CK:对照组,正常生长的中国沙棘幼苗;Pb_500、Pb_1000、Pb_2000、Pb_5000:实验组,铅胁迫浓度依次为500、1 000、2 000 和5 000 mg/kg。下同
Fig. 6 Expression analysis of NRAMP gene in H. rhamnoi-des under different concentrations of lead stress CK: Control group, normal-growing seabuckthorn seedlings. Pb_500, Pb_1000, Pb_2000, Pb_5000: Experimental groups, with Pb stress concentrations of 500, 1 000, 2 000, and 5 000 mg/kg. The same below
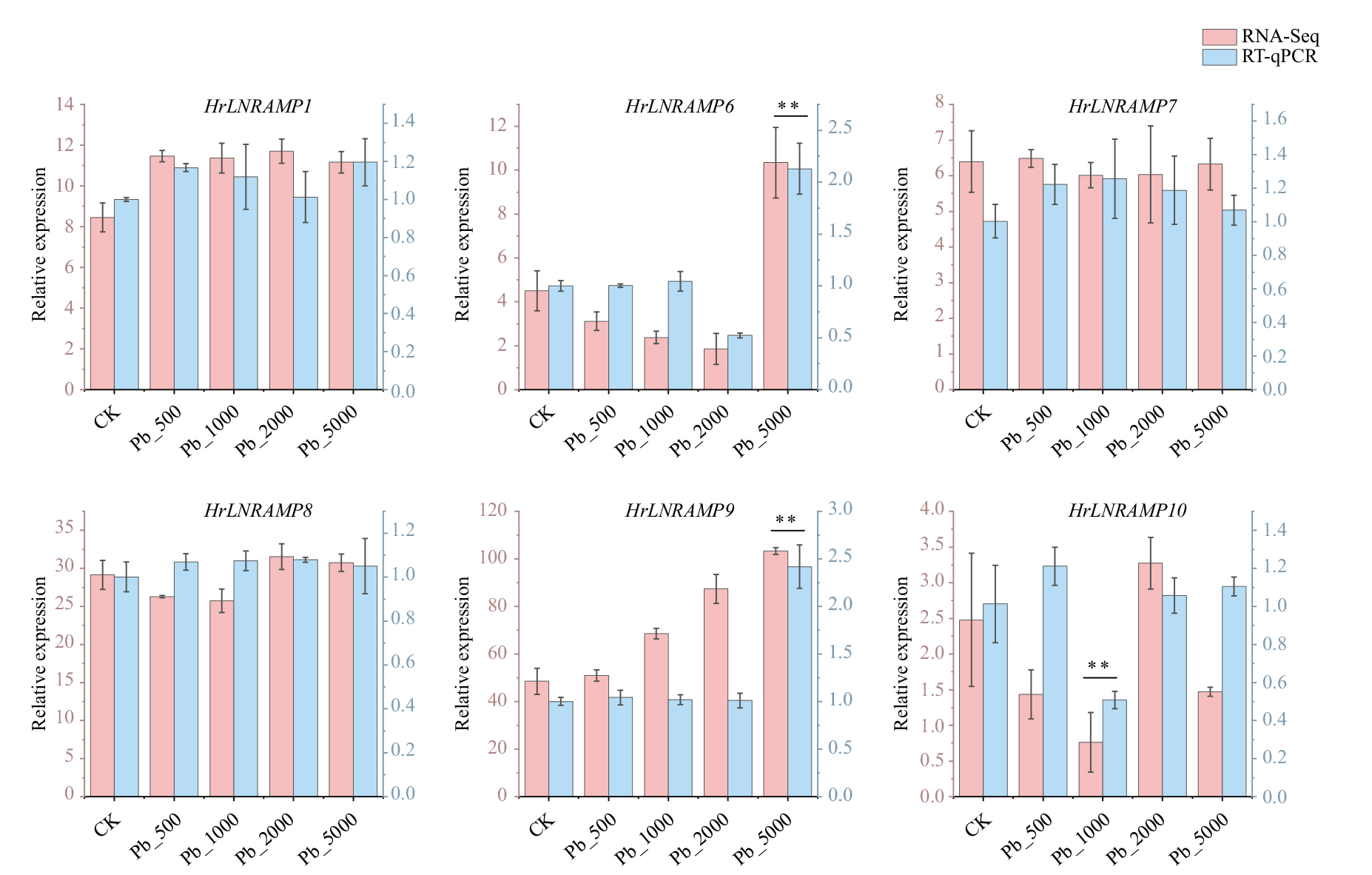
图7 铅胁迫条件下HrLNRAMP家族部分基因相对表达量 数据代表3个生物学重复的平均值,竖线表示标准差,星号表示相应基因在对照组和胁迫组之间差异显著(** P <0.01)
Fig. 7 Relative expressions of partial genes of HrLNRAMP gene family under lead stress conditions Data are means(±SD)of three biological replicates. Vertical bars indicate standard deviations. Asterisks indicate corresponding genes significantly upregulated or downregulated between the treatment and control(** P < 0.01)
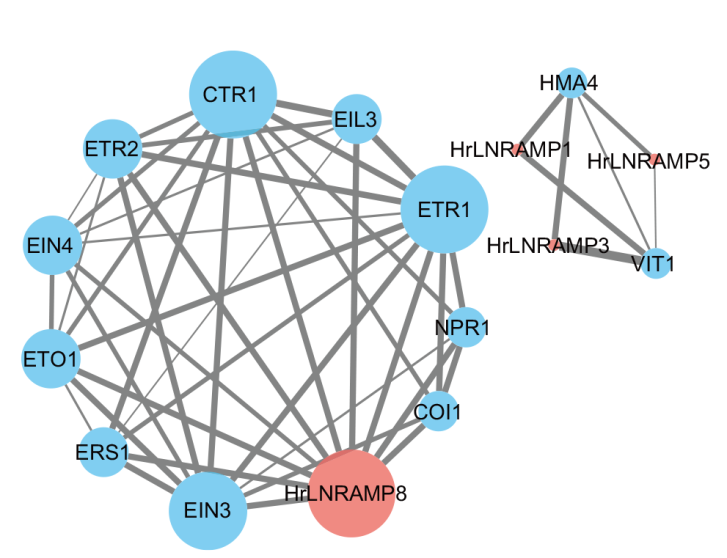
图10 沙棘NRAMP基因的蛋白互作网络 线条的粗细代表不同互作强度,线条粗,互作强度大,线条细,互作强度小。其中蓝色代表拟南芥中所有与沙棘存在互作关系的蛋白;红色仅代表存在互作关系的NRAMP蛋白
Fig. 10 Interaction protein network of HrLNRAMP geness in seabuckthorn Line thickness indicates the interaction strength; thicker lines denote stronger interactions, while thinner lines signify weaker ones. Blue denotes Arabidopsis proteins interacting with seabuckthorn, while red is specific to NRAMP proteins engaging in such interactions
| [1] | Nevo Y, Nelson N. The NRAMP family of metal-ion transporters[J]. Biochim Biophys Acta Mol Cell Res, 2006, 1763(7): 609-620. |
| [2] | Bozzi AT, Gaudet R. Molecular mechanism of NRAMP-family transition metal transport[J]. J Mol Biol, 2021, 433(16): 166991. |
| [3] |
Thomine S, Wang R, Ward JM, et al. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to NRAMP genes[J]. Proc Natl Acad Sci USA, 2000, 97(9): 4991-4996.
doi: 10.1073/pnas.97.9.4991 pmid: 10781110 |
| [4] | Gong ZH, Duan YQ, Liu DM, et al. Physiological and transcriptome analysis of response of soybean(Glycine max)to cadmium stress under elevated CO2 concentration[J]. J Hazard Mater, 2023, 448: 130950. |
| [5] | Tian WJ, He GD, Qin LJ, et al. Genome-wide analysis of the NRAMP gene family in potato(Solanum tuberosum): identification, expression analysis and response to five heavy metals stress[J]. Ecotoxicol Environ Saf, 2021, 208: 111661. |
| [6] | Fareed MM, Qasmi M, Shahid Z. The the structural, functional identification of natural resistance-associated macrophage protein as transporter elements in Oryza sativa[J]. European Journal of Volunteering and Community-based Projects, 2021, 1(2): 17-27. |
| [7] |
李一涵, 于浪柳, 李春燕, 等. 大麦NRAMP全基因组鉴定及重金属胁迫下基因表达分析[J]. 生物技术通报, 2022, 38(6): 103-111.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1006 |
| Li YH, Yu LL, Li CY, et al. Whole genome identification of barley NRAMP and gene expression analysis under heavy metal stress[J]. Biotechnol Bull, 2022, 38(6): 103-111. | |
| [8] | Hussain Q, Ye T, Shang CJ, et al. NRAMP gene family in Kandelia obovata: genome-wide identification, expression analysis, and response to five different copper stress conditions[J]. Front Plant Sci, 2024, 14: 1318383. |
| [9] | 陈可欣, 蒋贤达, 朱祝军, 等. 植物NRAMP家族参与金属离子吸收和分配的研究进展[J]. 植物生理学报, 2020, 56(3): 345-355. |
| Chen KX, Jiang XD, Zhu ZJ, et al. Advances in the study of plant NRAMP family involved in metal ion absorption and distribution[J]. Plant Physiol J, 2020, 56(3): 345-355. | |
| [10] | Kumar A, Singh G, Prasad B, et al. Genome wide analysis and identification of NRAMP gene family in wheat(Triticum aestivum L.)[J]. J Pharm Innov, 2022, 11(1): 499-504. |
| [11] |
Ihnatowicz A, Siwinska J, Meharg AA, et al. Conserved histidine of metal transporter AtNRAMP1 is crucial for optimal plant growth under manganese deficiency at chilling temperatures[J]. New Phytol, 2014, 202(4): 1173-1183.
doi: 10.1111/nph.12737 pmid: 24571269 |
| [12] | Żuchowski J. Phytochemistry and pharmacology of sea buckthorn(Elaeagnus rhamnoides; syn. Hippophae rhamnoides): progress from 2010 to 2021[J]. Phytochem Rev, 2023, 22(1): 3-33. |
| [13] | 徐智玮, 贾守宁, 赵国福, 等. 青海沙棘资源调查与产业发展建议[J]. 中国现代中药, 2020, 22(9): 1453-1457. |
| Xu ZW, Jia SN, Zhao GF, et al. Resources investigation and industrial development of Hippophae rhamnoides in Qinghai Province[J]. Mod Chin Med, 2020, 22(9): 1453-1457. | |
| [14] | Yu LY, Diao SF, Zhang GY, et al. Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides[J]. Plant Biotechnol J, 2022, 20(7): 1257-1273. |
| [15] |
Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis[J]. Plant Physiol, 2001, 126(4): 1646-1667.
doi: 10.1104/pp.126.4.1646 pmid: 11500563 |
| [16] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [17] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [18] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [19] | Bailey TL, Johnson J, Grant CE, et al. The MEME suite[J]. Nucleic Acids Res, 2015, 43(W1): W39-W49. |
| [20] |
Qin L, Han PP, Chen LY, et al. Genome-wide identification and expression analysis of NRAMP family genes in soybean(Glycine max L.)[J]. Front Plant Sci, 2017, 8: 1436.
doi: 10.3389/fpls.2017.01436 pmid: 28868061 |
| [21] | 杨猛. 水稻NRAMP家族基因在Mn和Cd转运中的功能研究[D]. 武汉: 华中农业大学, 2014. |
| Yang M. Functional analysis of rice NRAMP genes in Mn and Cd transport[D]. Wuhan: Huazhong Agricultural University, 2014. | |
| [22] | 陈娜. 沙棘修复尾矿坝的重金属迁移时间效应分析及预测[D]. 沈阳: 辽宁大学, 2021. |
| Chen N. Analysis and prediction of heavy metal migration time effect of sea buckthorn remediation tailings dam[D]. Shenyang: Liaoning University, 2021. | |
| [23] |
Wang PP, Moore BM, Panchy NL, et al. Factors influencing gene family size variation among related species in a plant family, Solanaceae[J]. Genome Biol Evol, 2018, 10(10): 2596-2613.
doi: 10.1093/gbe/evy193 pmid: 30239695 |
| [24] | Tian SK, Wang DD, Yang L, et al. A systematic review of 1-Deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants[J]. Plant Growth Regul, 2022, 96(2): 221-235. |
| [25] | Mani A, Sankaranarayanan K. In silico analysis of natural resistance-associated macrophage protein(NRAMP)family of transporters in rice[J]. Protein J, 2018, 37(3): 237-247. |
| [26] |
Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life[J]. Nat Rev Genet, 2005, 6(5): 361-375.
doi: 10.1038/nrg1603 pmid: 15861208 |
| [27] | Chang JD, Gao WP, Wang P, et al. OsNRAMP5 is a major transporter for lead uptake in rice[J]. Environ Sci Technol, 2022, 56(23): 17481-17490. |
| [28] |
Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology[J]. Nat Rev Genet, 2013, 14: 360-366.
doi: 10.1038/nrg3456 pmid: 23552219 |
| [29] | Tiwari M, Sharma D, Dwivedi S, et al. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance[J]. Plant Cell Environ, 2014, 37(1): 140-152. |
| [30] | Nie YY, Li Y, Liu MH, et al. The nucleoporin NUP160 and NUP96 regulate nucleocytoplasmic export of mRNAs and participate in ethylene signaling and response in Arabidopsis[J]. Plant Cell Rep, 2023, 42(3): 549-559. |
| [31] | Martin RE, Marzol E, Estevez JM, et al. Ethylene signaling increases reactive oxygen species accumulation to drive root hair initiation in Arabidopsis[J]. Development, 2022, 149(13): dev200487. |
| [32] | Gao HY, Xia XY, An LJ. Critical roles of the activation of ethylene pathway genes mediated by DNA demethylation in Arabidopsis hyperhydricity[J]. Plant Genome, 2022, 15(2): e20202. |
| [33] | Guedes FRCM, Maia CF, Silva BRSD, et al. Exogenous 24-Epibrassinolide stimulates root protection, and leaf antioxidant enzymes in lead stressed rice plants: central roles to minimize Pb content and oxidative stress[J]. Environ Pollut, 2021, 280: 116992. |
| [34] |
Thao NP, Khan MI, Thu NBA, et al. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress[J]. Plant Physiol, 2015, 169(1): 73-84.
doi: 10.1104/pp.15.00663 pmid: 26246451 |
| [35] | Zou Y, Ding XC, Ye LL, et al. Molecular cloning and expression analysis of EIN2, EIN3/EIL, and EBF genes during papaya fruit development and ripening[J]. N Z J Crop Hortic Sci, 2021, 49(2/3): 151-167. |
| [1] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [2] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [3] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [4] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [5] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [6] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [7] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [8] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| [9] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [10] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [11] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [12] | 谢宏, 周丽莹, 李舒文, 王梦迪, 艾晔, 晁跃辉. 蒺藜苜蓿MtCIM基因结构和功能分析[J]. 生物技术通报, 2024, 40(1): 262-269. |
| [13] | 唐伟林, 康琴, 汪霞, 谌明洋, 孙欣江, 王棵, 侯凯, 吴卫, 徐东北. 薄荷茉莉酸受体McCOI1a基因的克隆与表达模式分析[J]. 生物技术通报, 2024, 40(1): 270-280. |
| [14] | 江润海, 姜冉冉, 朱城强, 侯秀丽. 微生物强化植物修复铅污染土壤的机制研究进展[J]. 生物技术通报, 2023, 39(8): 114-125. |
| [15] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||