








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 131-140.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1163
郝思怡1( ), 张君珂1, 王斌2, 曲朋燕1, 李瑞得1, 程春振1(
), 张君珂1, 王斌2, 曲朋燕1, 李瑞得1, 程春振1( )
)
收稿日期:2023-12-07
出版日期:2024-05-26
发布日期:2024-06-13
通讯作者:
程春振,男,博士,副教授,研究方向:园艺植物生物技术;E-mail: ld0532cheng@126.com作者简介:郝思怡,女,硕士研究生,研究方向:果树生物技术;E-mail: yilia_fz@foxmail.com
基金资助:
HAO Si-yi1( ), ZHANG Jun-ke1, WANG Bin2, QU Peng-yan1, LI Rui-de1, CHENG Chun-zhen1(
), ZHANG Jun-ke1, WANG Bin2, QU Peng-yan1, LI Rui-de1, CHENG Chun-zhen1( )
)
Received:2023-12-07
Published:2024-05-26
Online:2024-06-13
摘要:
【目的】早花基因3(EARLY FLOWERING3, ELF3)是生物钟系统核心振荡器的重要组成成员,在植物生物钟和开花调控及逆境胁迫等过程扮演重要角色。目前尚无香蕉ELF3的相关报道,为揭示ELF3在香蕉抵御逆境胁迫中的功能对其进行了鉴定、克隆和表达分析。【方法】克隆了从香蕉基因组鉴定获得的4个MaELF3成员(MaELF3-1-MaELF3-4),利用生物信息学手段分析了它们的序列特征,基于转录组数据和实时荧光定量PCR研究了它们在高低温胁迫、香蕉枯萎病菌FocTR4侵染及茉莉酸甲酯(MeJA)和脱落酸(ABA)处理后的表达模式。【结果】4个MaELF3的CDS长度介于2 058-2 301 bp,可编码685-766 aa。除MaELF3-4含3个外显子外,其余MaELF3s均含4个外显子。4个MaELF3s均为定位于细胞核的不稳定碱性蛋白;与温带植物ELF3s不同,MaELF3s和多种热带植物的ELF3均无朊病毒样结构域(PrD)。系统进化结果显示,MaELF3-2和MaELF3-4与拟南芥ELF3(At2g25930)亲缘关系最近;MaELF3-1和MaELF3-3分别与粗柄象腿蕉(Ensete ventricosum)和野蕉(Musa balbisiana)ELF3亲缘关系最近。MaELF3s启动子上存在一些光、激素(MeJA、ABA等)和逆境胁迫(干旱、低温等)响应相关元件。基因表达分析结果显示,所有4个MaELF3s在香蕉叶片中的表达均受ABA和JA影响,且它们在香蕉根系中的表达受FocTR4显著抑制;除MaELF3-4外其他成员的表达均受高低温显著抑制。【结论】MaELF3s参与了香蕉对不同逆境胁迫的响应。
郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140.
HAO Si-yi, ZHANG Jun-ke, WANG Bin, QU Peng-yan, LI Rui-de, CHENG Chun-zhen. Cloning and Expression Analysis of Banana EARLY FLOWERING 3(ELF3)Genes[J]. Biotechnology Bulletin, 2024, 40(5): 131-140.
| 基因Gene | 引物名称Primer name | 引物序列Primer sequence(5'-3') | 退火温度Annealing temperature/℃ | 用途Application |
|---|---|---|---|---|
| MaELF3-1 | MaELF3-1-F | ATGAAAGGGGGAAAGGAGGA | 62 | 基因克隆 Gene cloning |
| MaELF3-1-R | TCAGTCATGTTGCAGCCTCTC | |||
| MaELF3-1-qF | ATCTACAGACGGAGAACATTGCT | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-1-qR | GCCCACTTTCTAAAACCTGACGATG | |||
| MaELF3-2 | MaELF3-2-F | ATGAAAGGGGCGGAGGAG | 62 | 基因克隆 Gene cloning |
| MaELF3-2-R | TCATGAGTCATGTTGTTGCCTC | |||
| MaELF3-2-qF | ATGCAATATGTCCCATCATCCGAA | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-2-qR | TTGCTGTGCTTCCTTGTAACTCG | |||
| MaELF3-3 | MaELF3-3-F | ATGAAAGGGGAGAAGGATGA | 58 | 基因克隆 Gene cloning |
| MaELF3-3-R | TCATGAATCATGCTGTTGTCTT | |||
| MaELF3-3-qF | ATGCAATATGTCCCATCATCCGAA | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-3-qR | TTGCTGTGCTTCCTTGTAACTCG | |||
| MaELF3-4 | MaELF3-4-F | ATGCTTTCTCCATTGTACATTTCTC | 60 | 基因克隆 Gene cloning |
| MaELF3-4-R | TCATGAGTCATGTTGTTGCCT | |||
| MaELF3-4-qF | ATCAACGAATGGCGATACAGT | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-4-qR | GCCTCACCATCATTAATAACAGC |
表1 所用引物信息
Table 1 Information of the primers used in this study
| 基因Gene | 引物名称Primer name | 引物序列Primer sequence(5'-3') | 退火温度Annealing temperature/℃ | 用途Application |
|---|---|---|---|---|
| MaELF3-1 | MaELF3-1-F | ATGAAAGGGGGAAAGGAGGA | 62 | 基因克隆 Gene cloning |
| MaELF3-1-R | TCAGTCATGTTGCAGCCTCTC | |||
| MaELF3-1-qF | ATCTACAGACGGAGAACATTGCT | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-1-qR | GCCCACTTTCTAAAACCTGACGATG | |||
| MaELF3-2 | MaELF3-2-F | ATGAAAGGGGCGGAGGAG | 62 | 基因克隆 Gene cloning |
| MaELF3-2-R | TCATGAGTCATGTTGTTGCCTC | |||
| MaELF3-2-qF | ATGCAATATGTCCCATCATCCGAA | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-2-qR | TTGCTGTGCTTCCTTGTAACTCG | |||
| MaELF3-3 | MaELF3-3-F | ATGAAAGGGGAGAAGGATGA | 58 | 基因克隆 Gene cloning |
| MaELF3-3-R | TCATGAATCATGCTGTTGTCTT | |||
| MaELF3-3-qF | ATGCAATATGTCCCATCATCCGAA | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-3-qR | TTGCTGTGCTTCCTTGTAACTCG | |||
| MaELF3-4 | MaELF3-4-F | ATGCTTTCTCCATTGTACATTTCTC | 60 | 基因克隆 Gene cloning |
| MaELF3-4-R | TCATGAGTCATGTTGTTGCCT | |||
| MaELF3-4-qF | ATCAACGAATGGCGATACAGT | 60 | 实时荧光定量PCR RT-qPCR | |
| MaELF3-4-qR | GCCTCACCATCATTAATAACAGC |
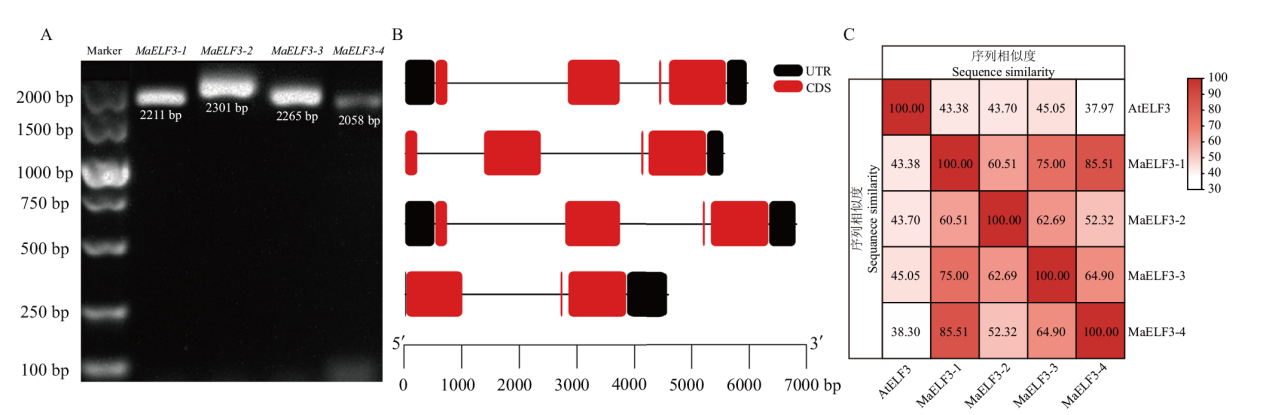
图1 MaELF3s的扩增(A)、基因结构(B)及编码蛋白相似度(C)的分析
Fig. 1 Analysis for the amplification of MaELF3s(A), gene structures(B), and sequence similarity of their encoded proteins(C)
| 蛋白 Protein | 基因ID Gene ID | 编码区长度 CDS length/bp | 蛋白长度 Protein size/aa | 分子量 Molecular weight/Da | 等电点 pI | 不稳定系数 Instability index | 最大平均亲水性 GRAVY | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| MaELF3-1 | Macma4_01_g08270 | 2 211 | 736 | 80 267.16 | 8.52 | 65.72 | -0.652 | 细胞核Nucleus |
| MaELF3-2 | Macma4_04_g26880 | 2 301 | 766 | 82 762.16 | 8.94 | 66.59 | -0.628 | 细胞核Nucleus |
| MaELF3-3 | Macma4_05_g09710 | 2 265 | 754 | 81 749.59 | 8.66 | 63.25 | -0.693 | 细胞核Nucleus |
| MaELF3-4 | Macma4_06_g37150 | 2 058 | 685 | 74 061.53 | 8.96 | 69.77 | -0.61 | 细胞核Nucleus |
表2 香蕉ELF3蛋白基本理化性质分析结果
Table 2 Basic physicochemical properties of the identified banana's ELF3 proteins
| 蛋白 Protein | 基因ID Gene ID | 编码区长度 CDS length/bp | 蛋白长度 Protein size/aa | 分子量 Molecular weight/Da | 等电点 pI | 不稳定系数 Instability index | 最大平均亲水性 GRAVY | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| MaELF3-1 | Macma4_01_g08270 | 2 211 | 736 | 80 267.16 | 8.52 | 65.72 | -0.652 | 细胞核Nucleus |
| MaELF3-2 | Macma4_04_g26880 | 2 301 | 766 | 82 762.16 | 8.94 | 66.59 | -0.628 | 细胞核Nucleus |
| MaELF3-3 | Macma4_05_g09710 | 2 265 | 754 | 81 749.59 | 8.66 | 63.25 | -0.693 | 细胞核Nucleus |
| MaELF3-4 | Macma4_06_g37150 | 2 058 | 685 | 74 061.53 | 8.96 | 69.77 | -0.61 | 细胞核Nucleus |

图2 香蕉ELF3蛋白序列比对结果 红色线条和绿色箭头分别代表α螺旋和β折叠
Fig. 2 Protein sequences alignment results of four banana ELF3s Red lines and green arrows indicate α-helix and β sheet, respectively
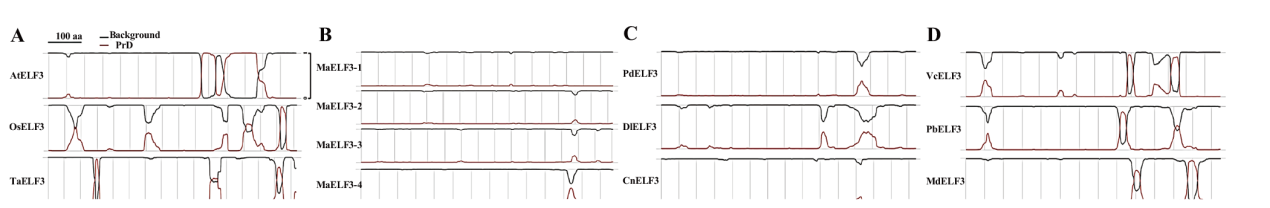
图3 不同植物ELF3蛋白PrD的预测 A:模式植物ELF3;B:香蕉ELF3;C:热带植物ELF3;D:温带植物ELF3。At:拟南芥;Os:水稻;Ta:小麦;Ma:香蕉;Pd:海枣;Dl:龙眼;Cn:椰子;Vc:蓝莓;Pb:梨;Md:苹果
Fig. 3 PrD domain prediction for ELF3 proteins from different plant species A: Model plant ELF3; B: banana ELF3; C: ELF3s from tropical plants; D: ELF3s from temperate plants. At: Arabidopsis thaliana; Os: Oryza sativa; Ta: Triticum aestivum;Ma: Musa acuminata; Pd: phoenix dactylifera; Dl: Dimocarpus longan; Cn: Cocos nucifera; Vc: Vaccinium corymbosum; pear Pyrus bretschneideri; Md: Malus domestica
| 功能Function | 元件Element | MaELF3-1 | MaELF3-2 | MaELF3-3 | MaELF3-4 | |
|---|---|---|---|---|---|---|
| 光响应 Light responsive | 光 Light | ACE | 1 | |||
| AE-box | 1 | 1 | ||||
| Box 4 | 1 | 8 | ||||
| Gap-box | 1 | |||||
| GATA-motif | 3 | 2 | ||||
| G-Box | 3 | |||||
| GT1-motif | 1 | 1 | 1 | |||
| GTGGC-motif | 1 | |||||
| I-box | 1 | 1 | 2 | |||
| Sp1 | 2 | 1 | ||||
| TCCC-motif | 1 | |||||
| TCT-motif | 2 | 1 | ||||
| 生长发育 Growth and development | 分生组织表达Meristem expression | CAT-box | 1 | 1 | 2 | 1 |
| 玉米蛋白代谢Zein metabolism | O2-site | 1 | ||||
| 激素响应 Phytohormone responsive | 脱落酸Abscisic acid | ABRE | 7 | 1 | 1 | |
| 生长素Auxin | AuxRR-core | 1 | ||||
| TGA-element | 1 | |||||
| 茉莉酸甲酯Methyl jasmonate | CGTCA-motif | 2 | 4 | 2 | 1 | |
| TGACG-motif | 2 | 4 | 2 | 1 | ||
| 赤霉素Gibberellin | GARE-motif | 1 | 1 | |||
| P-box | 1 | |||||
| TATC-box | 1 | |||||
| 水杨酸Salicylic acid | TCA-element | 1 | 1 | |||
| 胁迫响应 Stress responsive | 厌氧诱导Anaerobic induction | ARE | 1 | |||
| 缺氧特异性诱导Anoxic specific induction | GC-motif | 1 | 1 | 1 | 2 | |
| 低温Low temperature | LTR | 1 | ||||
| 干旱诱导Drought induction | MBS | 3 | 1 | |||
表3 MaELF3启动子顺式作用元件的预测
Table 3 Cis-acting elements predicted in the promoters of MaELF3s
| 功能Function | 元件Element | MaELF3-1 | MaELF3-2 | MaELF3-3 | MaELF3-4 | |
|---|---|---|---|---|---|---|
| 光响应 Light responsive | 光 Light | ACE | 1 | |||
| AE-box | 1 | 1 | ||||
| Box 4 | 1 | 8 | ||||
| Gap-box | 1 | |||||
| GATA-motif | 3 | 2 | ||||
| G-Box | 3 | |||||
| GT1-motif | 1 | 1 | 1 | |||
| GTGGC-motif | 1 | |||||
| I-box | 1 | 1 | 2 | |||
| Sp1 | 2 | 1 | ||||
| TCCC-motif | 1 | |||||
| TCT-motif | 2 | 1 | ||||
| 生长发育 Growth and development | 分生组织表达Meristem expression | CAT-box | 1 | 1 | 2 | 1 |
| 玉米蛋白代谢Zein metabolism | O2-site | 1 | ||||
| 激素响应 Phytohormone responsive | 脱落酸Abscisic acid | ABRE | 7 | 1 | 1 | |
| 生长素Auxin | AuxRR-core | 1 | ||||
| TGA-element | 1 | |||||
| 茉莉酸甲酯Methyl jasmonate | CGTCA-motif | 2 | 4 | 2 | 1 | |
| TGACG-motif | 2 | 4 | 2 | 1 | ||
| 赤霉素Gibberellin | GARE-motif | 1 | 1 | |||
| P-box | 1 | |||||
| TATC-box | 1 | |||||
| 水杨酸Salicylic acid | TCA-element | 1 | 1 | |||
| 胁迫响应 Stress responsive | 厌氧诱导Anaerobic induction | ARE | 1 | |||
| 缺氧特异性诱导Anoxic specific induction | GC-motif | 1 | 1 | 1 | 2 | |
| 低温Low temperature | LTR | 1 | ||||
| 干旱诱导Drought induction | MBS | 3 | 1 | |||
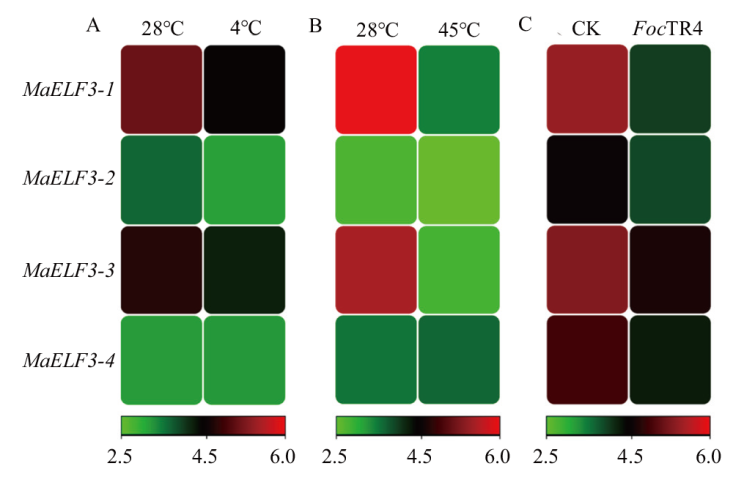
图6 MaELF3在低温(A)、高温处理(B)叶片以及FocTR4侵染根系(C)中的表达模式分析
Fig. 6 Expression patterns of MaELF3 genes in low(A)and high(B)temperature treated leaves, and in FocTR4 infected roots(C)
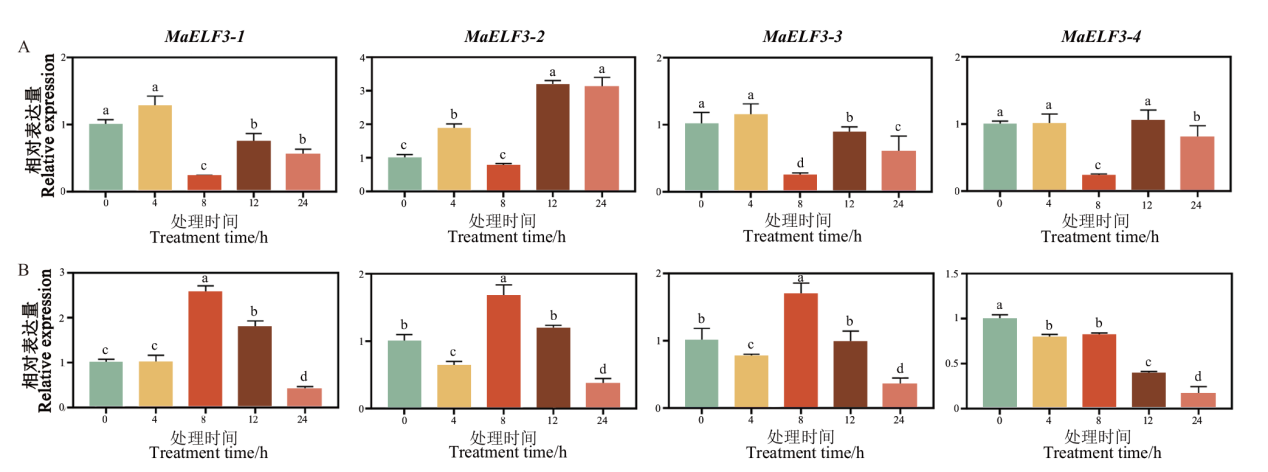
图7 MaELF3在ABA(A)和JA(B)处理后的表达模式 不同字母表示在P<0.05水平差异显著
Fig. 7 Expression patterns of MaELF3 genes in banana leaves at different times post ABA(A)and JA(B)treatments Different letters above columns indicate significant differences at P<0.05 level
| [1] | Creux N, Harmer S. Circadian rhythms in plants[J]. Cold Spring Harb Perspect Biol, 2019, 11(9): a034611. |
| [2] |
Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis[J]. Plant Cell, 2001, 13(6): 1281-1292.
pmid: 11402160 |
| [3] | Zagotta MT, Shannon S, Jacobs C, et al. Early-flowering mutants of Arabidopsis thaliana[J]. Funct Plant Biol, 1992, 19(4): 411. |
| [4] | Nusinow DA, Helfer A, Hamilton EE, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth[J]. Nature, 2011, 475(7356): 398-402. |
| [5] | Boden SA, Weiss D, Ross JJ, et al. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression[J]. Plant Cell, 2014, 26(4): 1557-1569. |
| [6] | Zhao H, Xu D, Tian T, et al. Molecular and functional dissection of EARLY-FLOWERING 3(ELF3)and ELF4 in Arabidopsis[J]. Plant Sci, 2021, 303: 110786. |
| [7] | Woods DP, Li WY, Sibout R, et al. PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon[J]. PLoS Genet, 2023, 19(5): e1010706. |
| [8] | Jung JH, Barbosa AD, Hutin S, et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis[J]. Nature, 2020, 585(7824): 256-260. |
| [9] | Hutin S, Kumita JR, Strotmann VI, et al. Phase separation and molecular ordering of the prion-like domain of the Arabidopsis thermosensory protein EARLY FLOWERING 3[J]. Proc Natl Acad Sci U S A, 2023, 120(28): e2304714120. |
| [10] | Box MS, Huang BE, Domijan M, et al. ELF3 controls thermoresponsive growth in Arabidopsis[J]. Curr Biol, 2015, 25(2): 194-199. |
| [11] |
Yang MK, Zhu XJ, Chen CM, et al. The plant circadian clock regulates autophagy rhythm through transcription factor LUX ARRHYTHMO[J]. J Integr Plant Biol, 2022, 64(11): 2135-2149.
doi: 10.1111/jipb.13343 |
| [12] |
Sun WJ, Han HY, Deng L, et al. Mediator subunit MED25 physically interacts with PHYTOCHROME INTERACTING FACTOR4 to regulate shade-induced hypocotyl elongation in tomato[J]. Plant Physiol, 2020, 184(3): 1549-1562.
doi: 10.1104/pp.20.00587 pmid: 33889988 |
| [13] | Zhang LL, Li W, Tian YY, et al. The E3 ligase XBAT35 mediates thermoresponsive hypocotyl growth by targeting ELF3 for degradation in Arabidopsis[J]. J Integr plant Biol, 2021, 63(6): 1097-1103. |
| [14] | Sakuraba Y, Jeong J, Kang MY, et al. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis[J]. Nat Commun, 2014, 5: 4636. |
| [15] | Sakuraba Y, Bülbül S, Piao WL, et al. Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways[J]. Plant J, 2017, 92(6): 1106-1120. |
| [16] |
Devlin PF, Robson PR, Patel SR, et al. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time[J]. Plant Physiol, 1999, 119(3): 909-915.
pmid: 10069829 |
| [17] |
Mizoguchi T, Wright L, Fujiwara S, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis[J]. Plant Cell, 2005, 17(8): 2255-2270.
doi: 10.1105/tpc.105.033464 pmid: 16006578 |
| [18] | Jiang YP, Yang CW, Huang S, et al. The ELF3-PIF7 interaction mediates the circadian gating of the shade response in Arabidopsis[J]. iScience, 2019, 22: 288-298. |
| [19] |
Pereira A, Maraschin M. Banana(Musa spp)from peel to pulp ethnopharmacology, source of bioactive compounds and its relevance for human health[J]. J Ethnopharmacol, 2015, 160: 149-163.
doi: 10.1016/j.jep.2014.11.008 pmid: 25449450 |
| [20] | Bodjrenou DM, Cheng CZ, Sun XL, et al. High temperature associated microRNAs and their potential roles in mediating heat tolerance in the leaf of banana inoculated with Serendipita indica[J]. J Hortic Sci Biotechnol, 2022, 97(2): 171-186. |
| [21] | Siamak SB, Zheng SJ. Banana Fusarium wilt(Fusarium oxysporum f. sp. cubense)control and resistance, in the context of developing wilt-resistant bananas within sustainable production systems[J]. Hortic Plant J, 2018, 4(5): 208-218. |
| [22] | 甘林, 代玉立, 刘晓菲, 等. 香蕉枯萎病高效拮抗土著细菌的筛选及其防效[J]. 西北农林科技大学学报:自然科学版, 2023. DOI: 10.13207/j.cnki.jnwafu.2024.06.010. |
| Gan L, Dai YL, Liu XF, et al. Screening and control of banana wilt disease in a highly effective antagonistic manner against indigenous bacteria[J]. J Northwest A & F Univ Nat Sci Ed, 2023. DOI: 10.13207/j.cnki.jnwafu.2024.06.010. | |
| [23] |
孙雪丽, 刘范, 田娜, 等. 香蕉Aux/IAA基因家族的全基因组鉴定及表达分析[J]. 园艺学报, 2019, 46(10): 1919-1935.
doi: 10.16420/j.issn.0513-353x.2018-0743 |
| Sun XL, Liu F, Tian N, et al. Genome-wide identification and expression analysis of Aux/IAA gene family in banana[J]. Acta Hortic Sin, 2019, 46(10): 1919-1935. | |
| [24] | 刘范, 田娜, 孙雪丽, 等. 香蕉GLP基因家族全基因组鉴定及表达分析[J]. 园艺学报, 2020, 47(10): 1930-1946. |
| Liu F, Tian N, Sun XL, et al. Genome-wide identification and expression analysis of banana GLP gene family[J]. Acta Hortic Sin, 2020, 47(10): 1930-1946. | |
| [25] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: A “One for All, All for One” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [26] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [27] |
Liu XL, Covington MF, Fankhauser C, et al. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway[J]. Plant Cell, 2001, 13(6): 1293-1304.
pmid: 11402161 |
| [28] |
Nieto C, López-Salmerón V, Davière JM, et al. ELF3-PIF4 interaction regulates plant growth independently of the evening complex[J]. Curr Biol, 2015, 25(2): 187-193.
doi: S0960-9822(14)01418-3 pmid: 25557667 |
| [29] | Xiang SY, Wu SG, Zhang HY, et al. The PIFs redundantly control plant defense response against Botrytis cinerea in Arabidopsis[J]. Plants, 2020, 9(9): 1246. |
| [30] | Cannon S, Kay W, Kilaru S, et al. Multi-site fungicides suppress banana Panama disease, caused by Fusarium oxysporum f. sp. cubense Tropical Race 4[J]. PLoS Pathog, 2022, 18(10): e1010860. |
| [31] | Zhao X, Huang LJ, Sun XF, et al. Transcriptomic and metabolomic analyses reveal key metabolites, pathways and candidate genes in Sophora davidii(franch.) skeels seedlings under drought stress[J]. Front Plant Sci, 2022, 13: 785702. |
| [32] | Zheng YY, Wang N, Zhang ZY, et al. Identification of flowering regulatory networks and hub genes expressed in the leaves of Elymus sibiricus L. using comparative transcriptome analysis[J]. Front Plant Sci, 2022, 13: 877908. |
| [33] |
Major IT, Yoshida Y, Campos ML, et al. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN(JAZ)-MYC transcriptional module[J]. New Phytol, 2017, 215(4): 1533-1547.
doi: 10.1111/nph.14638 pmid: 28649719 |
| [34] | Ye NH, Jia LG, Zhang JH. ABA signal in rice under stress conditions[J]. Rice, 2012, 5(1): 1. |
| [35] | Wang X, Zhang J, Song J, et al. Abscisic acid and hydrogen peroxide are involved in drought priming-induced drought tolerance in wheat(Triticum aestivum L.)[J]. Plant Biol, 2020, 22(6): 1113-1122. |
| [1] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [2] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [3] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [4] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [5] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [6] | 朱毅, 柳唐镜, 宫国义, 张洁, 王晋芳, 张海英. 西瓜ClPP2C3克隆及表达分析[J]. 生物技术通报, 2024, 40(1): 243-249. |
| [7] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [8] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [9] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [10] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [11] | 滕梦鑫, 徐亚, 何静, 汪奇, 乔飞, 李敬阳, 李新国. 香蕉MaMC6的克隆及原核表达分析[J]. 生物技术通报, 2023, 39(12): 179-186. |
| [12] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [13] | 侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135. |
| [14] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [15] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||