








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 223-232.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0989
任延靖1,2( ), 张鲁刚3, 赵孟良1, 李江1,2, 邵登魁1,2(
), 张鲁刚3, 赵孟良1, 李江1,2, 邵登魁1,2( )
)
收稿日期:2023-10-20
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
邵登魁,男,博士,研究员,硕士生导师,研究方向:蔬菜遗传育种;E-mail: 2006990015@qhu.edu.cn作者简介:任延靖,女,博士,副研究员,硕士生导师,研究方向:蔬菜遗传育种与分子生物学;E-mail: renyan0202@163.com
基金资助:
REN Yan-jing1,2( ), ZHANG Lu-gang3, ZHAO Meng-liang1, LI Jiang1,2, SHAO Deng-kui1,2(
), ZHANG Lu-gang3, ZHAO Meng-liang1, LI Jiang1,2, SHAO Deng-kui1,2( )
)
Received:2023-10-20
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】通过构建白菜种子的cDNA文库,筛选WDR 40蛋白TRANSPARENT TESTA GLABRA 1(TTG1)的互作蛋白,探究TTG1参与MBW三元复合体调控种皮原花青素形成的分子机制。【方法】以棕籽白菜自交系‘B147’的种子为材料,提取总RNA并建立cDNA文库,通过gateway技术构建诱饵载体pGBKT7-TTG1并进行酵母双杂交筛库。【结果】酵母文库库容为1.2×107 CFU,文库滴度是5.0×107 CFU/mL,插入片段平均长度大于1 000 bp,诱饵载体在酵母中无自激活活性。通过构建的诱饵载体pGBKT7-TTG1与构建的cDNA文库杂交,共获得了38个阳性互作蛋白,功能预测显示其中一个蛋白注释为MYB转录因子,注释为MYB73,序列分析结果显示该基因含有R2R3-MYB型抑制子保守基序C1和C2,推测该基因为白菜中参与种皮颜色形成的R2R3-MYB型抑制子,暗示着白菜中可能存在不同MYB转录因子参与的调控网络,影响着原花青素的形成。【结论】本研究构建了白菜种子组织的酵母双杂交cDNA文库,获得了38个TTG1阳性互作蛋白,首次挖掘到了可能影响白菜种皮颜色原花青素形成的R2R3-MYB型抑制子MYB73,为后期探究白菜种皮原花青素的调控网络奠定良好的基础。
任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232.
REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins[J]. Biotechnology Bulletin, 2024, 40(2): 223-232.
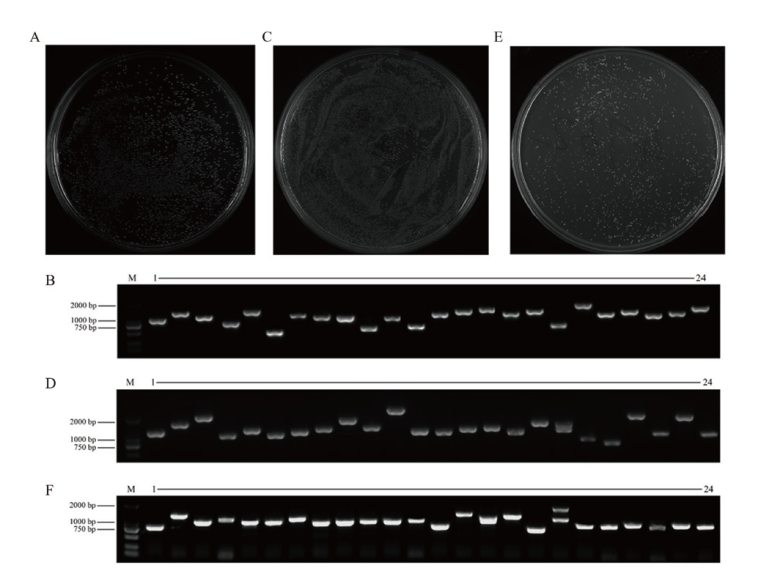
图1 文库的构建及鉴定 A:初级文库库容鉴定;B:初级文库插入片段鉴定;C:次级文库库容鉴定;D:次级文库插入片段鉴定;E:酵母文库库容鉴定;F:酵母文库插入片段鉴定
Fig. 1 Construction and identification of library A: Identification of the capacity of the primary library. B: Identification of inserts in the primary library. C: Identification of the capacity of the secondary library.D: Identification of inserts in the secondary library. E: Identification of the capacity of the yeast two-hybrid library. F: Identification of inserts in the yeast two-hybrid library

图2 pGBKT7-TTG1诱饵载体的自激活鉴定 A:阳性对照;B:阴性对照;C:SD/-Leu/-Trp/X-α-gal培养基生长情况;D:SD/-Leu/-Trp/-His/X-α-gal培养基生长情况;E:SD/-Leu/-Trp/-His/-Ade/X-α-gal/AbA培养基上生长情况
Fig. 2 Self-activation identification of pGBKT7-TTG1 bait vector A: Positive control. B: Negative control. C: SD/-Leu/-Trp/X-α-gal growth of gal medium. D: SD/-Leu/-Trp/X-α-gal growth of gal medium. E: SD/-Leu/-Trp/X-α-gal growth on gal/AbA medium

图3 BrTTG1在SD/-Leu/-Trp/-His/-Ade/X-α-gal/AbA(QDO/X/A)平板筛选的蓝色克隆 “-”为阴性对照;“+”为阳性对照
Fig. 3 Blue colonies screened by BrTTG1 in SD/-Leu/-Trp/-His/-Ade/X-α-gal/AbA(QDO/X/A)medium “-” represent negative control; “+” represent positive control
| 克隆编号Clone No. | 基因注释Gene annotation | 登录号GenBank access No. | 基因ID Gene ID |
|---|---|---|---|
| 1 | Lipoamide dehydrogenase precursor, partial[Arabidopsis thaliana] | BAD95420.1 | BraA06g004870.3C |
| 2 | Hypothetical protein BRARA_C02124[Brassica rapa] | RID70072.1 | BraA03g021980.3C |
| 3 | Multiple organellar RNA editing factor 2, chloroplastic[B. rapa] | XP_009143860.1 | BraA05g011400.3C |
| 5 | Transcription factor MYB44[B. napus] | NP_001303071.1 | BraA01g001590.3C |
| 6 | DNA polymerase zeta processivity subunit[B. rapa] | XP_009117755.1 | BraA09g058190.3C |
| 7 | SNF1-related protein kinase regulatory subunit beta-2 isoform X1[B. napus] | XP_013655743.1 | BraA08g011810.3C |
| 10 | Protein phosphatase 2C 37[B. rapa] | XP_009146731.2 | BraA05g035520.3C |
| 12 | Hypothetical protein F2Q68_00033308, partial[B. cretica] | KAF2544242.1 | BraA03g030840.3C |
| 13 | ADP, ATP carrier protein 2, mitochondrial[B. rapa] | XP_009131345.2 | BraA03g005780.3C |
| 17 | Hypothetical protein F2Q69_00061138, partial[B. cretica] | KAF3574004.1 | BraA07g015680.3C |
| 24 | Hypothetical protein BRARA_I05150[B. rapa] | RID48647.1 | BraA09g061120.3C |
| 25 | Hypothetical protein F2Q70_00009385[B. cretica] | KAF2614836.1 | BraA09g001710.3C |
| 27 | Hypothetical protein BRARA_A02089[B. rapa] | RID79344.1 | BraA01g022400.3C |
| 31 | Unnamed protein product[B. rapa] | VDC79045.1 | BraA03g009070.3C |
| 33 | BnaA01g23700D[B. napus] | CDY18109.1 | BraA01g030470.3C |
| 39 | Unnamed protein product[B. oleracea] | VDD24561.1 | BraA02g028520.3C |
| 40 | Scarecrow-like protein 8[B. rapa] | XP_009127236.1 | BraA02g015300.3C |
| 43 | LOW QUALITY PROTEIN: tripeptidyl-peptidase 2-like[B. rapa] | XP_009137154.1 | BraA01g011620.3C |
| 44 | BnaC08g42880D[B. napus] | CDY22654.1 | BraA09g062530.3C |
| 45 | BnaC03g15020D[B. napus] | CDY38747.1 | BraA03g014440.3C |
| 46 | Transcription initiation factor IIE subunit beta isoform X2[B. rapa] | XP_009128687.1 | BraA02g028520.3C |
| 47 | Hypothetical protein BRARA_F02580[B. rapa] | RID59345.1 | BraA06g030810.3C |
| 48 | Hypothetical protein BRARA_G03388[B. rapa] | RID56172.1 | BraA07g039370.3C |
| 50-53 | COP9 signalosome complex subunit 5a[B. rapa] | XP_009115580.1 | BraA09g040180.3C |
| 54 | Thioredoxin M2, chloroplastic-like isoform X1[B. napus] | XP_013642642.1 | BraA09g002600.3C |
| 55 | E3 ubiquitin-protein ligase RGLG2[B. rapa] | XP_009121662.1 | BraA10g024620.3C |
| 56 | Uncharacterized protein LOC103841884[B. rapa] | XP_009116717.1 | BraA09g050150.3C |
| 57 | Hypothetical protein BRARA_E01984[B. rapa] | RID62948.1 | BraA05g024640.3C |
| 59 | Probable pectin methylesterase CGR2[B. rapa] | XP_009149708.1 | BraA06g017430.3C |
| 62 | E3 ubiquitin-protein ligase MBR2[B. rapa] | XP_009104796.1 | BraA07g028570.3C |
| 64 | Chitinase-like protein 1 isoform X1[B. rapa] | XP_009111023.1 | BraA08g034170.3C |
| 65 | GDSL esterase/lipase At1g71250, partial[B. rapa] | XP_009105797.3 | BraA07g035870.3C |
| 67 | PREDICTED: aconitate hydratase 1-like[Raphanus sativus] | XP_018463972.1 | BraA08g020550.3C |
| 68 | Hypothetical protein BRARA_J00468[B. rapa] | RID40424.1 | BraA10g005490.3C |
| 70 | NADPH-dependent oxidoreductase 2-alkenal reductase[B. rapa] | XP_009130330.1 | BraA05g041630.3C |
| 73 | SNF1-related protein kinase regulatory subunit beta-2 isoform X1[B. napus] | XP_013655743.1 | BraA08g011810.3C |
| 76 | Catalase-3 isoform X1[B. rapa] | XP_009149539.1 | BraA07g015680.3C |
| 82 | Mitogen-activated protein kinase 7[B. napus] | NP_001303162.1 | BraA07g002560.3C |
表1 pGBKT7-TTG1互作蛋白注释
Table 1 pGBKT7-TTG1 interaction protein annotation
| 克隆编号Clone No. | 基因注释Gene annotation | 登录号GenBank access No. | 基因ID Gene ID |
|---|---|---|---|
| 1 | Lipoamide dehydrogenase precursor, partial[Arabidopsis thaliana] | BAD95420.1 | BraA06g004870.3C |
| 2 | Hypothetical protein BRARA_C02124[Brassica rapa] | RID70072.1 | BraA03g021980.3C |
| 3 | Multiple organellar RNA editing factor 2, chloroplastic[B. rapa] | XP_009143860.1 | BraA05g011400.3C |
| 5 | Transcription factor MYB44[B. napus] | NP_001303071.1 | BraA01g001590.3C |
| 6 | DNA polymerase zeta processivity subunit[B. rapa] | XP_009117755.1 | BraA09g058190.3C |
| 7 | SNF1-related protein kinase regulatory subunit beta-2 isoform X1[B. napus] | XP_013655743.1 | BraA08g011810.3C |
| 10 | Protein phosphatase 2C 37[B. rapa] | XP_009146731.2 | BraA05g035520.3C |
| 12 | Hypothetical protein F2Q68_00033308, partial[B. cretica] | KAF2544242.1 | BraA03g030840.3C |
| 13 | ADP, ATP carrier protein 2, mitochondrial[B. rapa] | XP_009131345.2 | BraA03g005780.3C |
| 17 | Hypothetical protein F2Q69_00061138, partial[B. cretica] | KAF3574004.1 | BraA07g015680.3C |
| 24 | Hypothetical protein BRARA_I05150[B. rapa] | RID48647.1 | BraA09g061120.3C |
| 25 | Hypothetical protein F2Q70_00009385[B. cretica] | KAF2614836.1 | BraA09g001710.3C |
| 27 | Hypothetical protein BRARA_A02089[B. rapa] | RID79344.1 | BraA01g022400.3C |
| 31 | Unnamed protein product[B. rapa] | VDC79045.1 | BraA03g009070.3C |
| 33 | BnaA01g23700D[B. napus] | CDY18109.1 | BraA01g030470.3C |
| 39 | Unnamed protein product[B. oleracea] | VDD24561.1 | BraA02g028520.3C |
| 40 | Scarecrow-like protein 8[B. rapa] | XP_009127236.1 | BraA02g015300.3C |
| 43 | LOW QUALITY PROTEIN: tripeptidyl-peptidase 2-like[B. rapa] | XP_009137154.1 | BraA01g011620.3C |
| 44 | BnaC08g42880D[B. napus] | CDY22654.1 | BraA09g062530.3C |
| 45 | BnaC03g15020D[B. napus] | CDY38747.1 | BraA03g014440.3C |
| 46 | Transcription initiation factor IIE subunit beta isoform X2[B. rapa] | XP_009128687.1 | BraA02g028520.3C |
| 47 | Hypothetical protein BRARA_F02580[B. rapa] | RID59345.1 | BraA06g030810.3C |
| 48 | Hypothetical protein BRARA_G03388[B. rapa] | RID56172.1 | BraA07g039370.3C |
| 50-53 | COP9 signalosome complex subunit 5a[B. rapa] | XP_009115580.1 | BraA09g040180.3C |
| 54 | Thioredoxin M2, chloroplastic-like isoform X1[B. napus] | XP_013642642.1 | BraA09g002600.3C |
| 55 | E3 ubiquitin-protein ligase RGLG2[B. rapa] | XP_009121662.1 | BraA10g024620.3C |
| 56 | Uncharacterized protein LOC103841884[B. rapa] | XP_009116717.1 | BraA09g050150.3C |
| 57 | Hypothetical protein BRARA_E01984[B. rapa] | RID62948.1 | BraA05g024640.3C |
| 59 | Probable pectin methylesterase CGR2[B. rapa] | XP_009149708.1 | BraA06g017430.3C |
| 62 | E3 ubiquitin-protein ligase MBR2[B. rapa] | XP_009104796.1 | BraA07g028570.3C |
| 64 | Chitinase-like protein 1 isoform X1[B. rapa] | XP_009111023.1 | BraA08g034170.3C |
| 65 | GDSL esterase/lipase At1g71250, partial[B. rapa] | XP_009105797.3 | BraA07g035870.3C |
| 67 | PREDICTED: aconitate hydratase 1-like[Raphanus sativus] | XP_018463972.1 | BraA08g020550.3C |
| 68 | Hypothetical protein BRARA_J00468[B. rapa] | RID40424.1 | BraA10g005490.3C |
| 70 | NADPH-dependent oxidoreductase 2-alkenal reductase[B. rapa] | XP_009130330.1 | BraA05g041630.3C |
| 73 | SNF1-related protein kinase regulatory subunit beta-2 isoform X1[B. napus] | XP_013655743.1 | BraA08g011810.3C |
| 76 | Catalase-3 isoform X1[B. rapa] | XP_009149539.1 | BraA07g015680.3C |
| 82 | Mitogen-activated protein kinase 7[B. napus] | NP_001303162.1 | BraA07g002560.3C |
| [1] |
Zhang JF, Lu Y, Yuan YX, et al. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa[J]. Plant Mol Biol, 2009, 69(5): 553-563.
doi: 10.1007/s11103-008-9437-y URL |
| [2] |
Ren YJ, Zhang N, Li R, et al. Comparative transcriptome and flavonoids components analysis reveal the structural genes responsible for the yellow seed coat color of Brassica rapa L[J]. PeerJ, 2021, 9: e10770.
doi: 10.7717/peerj.10770 URL |
| [3] | 任延靖. 白菜种子黄色种皮形成的分子机理[D]. 杨凌: 西北农林科技大学, 2018. |
| Ren YJ. Molecular mechanism of yellow seed coat formation in Chinese cabbage seeds[D]. Yangling: Northwest A & F University, 2018. | |
| [4] |
Yu CY. Molecular mechanism of manipulating seed coat coloration in oilseed Brassica species[J]. J Appl Genet, 2013, 54(2): 135-145.
doi: 10.1007/s13353-012-0132-y URL |
| [5] |
Lepiniec L, Debeaujon I, Routaboul JM, et al. Genetics and biochemistry of seed flavonoids[J]. Annu Rev Plant Biol, 2006, 57: 405-430.
pmid: 16669768 |
| [6] |
Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity[J]. Trends Plant Sci, 2005, 10(2): 63-70.
doi: 10.1016/j.tplants.2004.12.011 pmid: 15708343 |
| [7] |
Smith TF, Gaitatzes C, Saxena K, et al. The WD repeat: a common architecture for diverse functions[J]. Trends Biochem Sci, 1999, 24(5): 181-185.
pmid: 10322433 |
| [8] |
Walker AR, Davison PA, Bolognesi-Winfield AC, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein[J]. Plant Cell, 1999, 11(7): 1337-1350.
doi: 10.1105/tpc.11.7.1337 pmid: 10402433 |
| [9] |
Szymanski DB, Lloyd AM, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis[J]. Trends Plant Sci, 2000, 5(5): 214-219.
doi: 10.1016/s1360-1385(00)01597-1 pmid: 10785667 |
| [10] |
Baudry A, Heim MA, Dubreucq B, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana[J]. Plant J, 2004, 39(3): 366-380.
doi: 10.1111/tpj.2004.39.issue-3 URL |
| [11] |
Xu WJ, Lepiniec L, Dubos C. New insights toward the transcriptional engineering of proanthocyanidin biosynthesis[J]. Plant Signal Behav, 2014, 9(4): e28736.
doi: 10.4161/psb.28736 URL |
| [12] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [13] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112: 531-552.
pmid: 10027275 |
| [14] | Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Comput Appl Biosci, 1995, 11(6): 681-684. |
| [15] |
Fields S, Song O. A novel genetic system to detect protein-protein interactions[J]. Nature, 1989, 340(6230): 245-246.
doi: 10.1038/340245a0 |
| [16] |
Imamura T, Yasui YS, Koga H, et al. A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa[J]. Commun Biol, 2020, 3(1): 513.
doi: 10.1038/s42003-020-01249-w pmid: 32943738 |
| [17] |
Liu CG, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula[J]. Plant Physiol, 2014, 165(4): 1424-1439.
doi: 10.1104/pp.114.241877 URL |
| [18] |
Zhu QL, Sui SZ, Lei XH, et al. Ectopic expression of the Coleus R2R3 MYB-type proanthocyanidin regulator gene SsMYB3 alters the flower color in transgenic tobacco[J]. PLoS One, 2015, 10(10): e0139392.
doi: 10.1371/journal.pone.0139392 URL |
| [19] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [20] |
Zhou H, Lin-Wang K, Wang FR, et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation[J]. New Phytol, 2019, 221(4): 1919-1934.
doi: 10.1111/nph.15486 pmid: 30222199 |
| [21] |
Rajput R, Naik J, Stracke R, et al. Interplay between R2R3 MYB-type activators and repressors regulates proanthocyanidin biosynthesis in banana(Musa acuminata)[J]. New Phytol, 2022, 236(3): 1108-1127.
doi: 10.1111/nph.v236.3 URL |
| [22] |
Qu CM, Zhu MC, Hu R, et al. Comparative genomic analyses reveal the genetic basis of the yellow-seed trait in Brassica napus[J]. Nat Commun, 2023, 14(1): 5194.
doi: 10.1038/s41467-023-40838-1 |
| [1] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [2] | 辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211. |
| [3] | 朱毅, 柳唐镜, 宫国义, 张洁, 王晋芳, 张海英. 西瓜ClPP2C3克隆及表达分析[J]. 生物技术通报, 2024, 40(1): 243-249. |
| [4] | 谢宏, 周丽莹, 李舒文, 王梦迪, 艾晔, 晁跃辉. 蒺藜苜蓿MtCIM基因结构和功能分析[J]. 生物技术通报, 2024, 40(1): 262-269. |
| [5] | 唐伟林, 康琴, 汪霞, 谌明洋, 孙欣江, 王棵, 侯凯, 吴卫, 徐东北. 薄荷茉莉酸受体McCOI1a基因的克隆与表达模式分析[J]. 生物技术通报, 2024, 40(1): 270-280. |
| [6] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [7] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [8] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [9] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [10] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [11] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [12] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [13] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [14] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [15] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||