








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 64-73.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0133
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
收稿日期:2024-02-02
出版日期:2024-09-26
发布日期:2024-06-26
通讯作者:
高雅彬,女,硕士,副教授,研究方向:园艺植物生物技术、环境保护及资源再生利用;E-mail: lzbwkjxy_gyb@126.com作者简介:申鹏,男,硕士,讲师,研究方向:园艺植物生物技术、植物修复和栽培生理;E-mail: lzbwkjxy_sp@126.com
基金资助:
SHEN Peng( ), GAO Ya-Bin(
), GAO Ya-Bin( ), DING Hong
), DING Hong
Received:2024-02-02
Published:2024-09-26
Online:2024-06-26
摘要:
【目的】丝氨酸乙酰转移酶(SAT)是硫同化为半胱氨酸(cysteine, Cys)的关键酶,参与植物多种生物过程,特别是在植物响应非生物胁迫中发挥着重要作用。但关于马铃薯SAT基因家族(StSAT)的分析尚未见报道。系统鉴定了马铃薯SAT基因家族,为深入了解StSAT基因家族的特征,进一步分析它们在马铃薯抵御非生物胁迫中的功能提供了理论依据。【方法】利用HMM 对马铃薯SAT基因家族进行鉴定,并对其染色体分布、基因结构、蛋白保守基序及物种间的共线性进行分析。利用PGSC下载的RNA-seq数据分析双单倍体(doubled-monoploid, DM)马铃薯中StSATs在不同组织部位、非生物胁迫和外源激素处理下的表达模式。通过qPCR(quantitative real-time PCR)分析四倍体马铃薯中StSATs在NaCl和PEG处理(0、1、3和24 h)下的相对表达水平。【结果】在马铃薯中鉴定出4个StSATs,它们分布在4条染色体上。根据系统发育特征,将4个StSATs分在3个亚族中。共线性分析发现,StSATs与拟南芥(Arabidopsis thaliana)、番茄(Solanum lycopersicum)、甘蓝(Brassica oleracea)、水稻(Oryza sativa)和玉米(Zea mays)中分别有4对、4对、2对、1对和1对直系同源基因。通过表达分析发现,四倍体马铃薯中4个StSATs随着NaCl和PEG处理时间的延长,其表达量显著升高(与0 h相比),很可能参与马铃薯对盐和渗透胁迫的响应。【结论】StSAT基因家族成员在马铃薯响应盐和渗透胁迫中发挥着重要作用。
申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73.
SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.)[J]. Biotechnology Bulletin, 2024, 40(9): 64-73.
| 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Soltu06G005680 | GGTCGTGTTGAGACTGGTGTGATC | GCTTCGTGGTGCATCTCTACAGAC |
| Soltu02G022850 | CTGTCGCTAACACCGCTTCTACTC | GCTCAATGCCCGTTCCAAACAATC |
| Soltu12G025770 | GACACGGCGGAAGAAGCTATCTG | AGTGAGCGTTCAAGCGAAGAGTG |
| Soltu07G027060 | CCAAACAAGCCCCAAATCGACAAC | GCAGGAAGCAAGGTCAGGGAAAG |
| Soltu10G001180 | TGGTGCTGGTACTTGTGTTCTTGG | AGCAGTAGTTCTTGCAGGCACTTC |
表1 qPCR引物信息
Table 1 Primer information for qPCR
| 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Soltu06G005680 | GGTCGTGTTGAGACTGGTGTGATC | GCTTCGTGGTGCATCTCTACAGAC |
| Soltu02G022850 | CTGTCGCTAACACCGCTTCTACTC | GCTCAATGCCCGTTCCAAACAATC |
| Soltu12G025770 | GACACGGCGGAAGAAGCTATCTG | AGTGAGCGTTCAAGCGAAGAGTG |
| Soltu07G027060 | CCAAACAAGCCCCAAATCGACAAC | GCAGGAAGCAAGGTCAGGGAAAG |
| Soltu10G001180 | TGGTGCTGGTACTTGTGTTCTTGG | AGCAGTAGTTCTTGCAGGCACTTC |
| 基因ID Gene ID | 染色体定位 Chromosome localization | 氨基酸长度 Amino acid length | 相对分子量 Molecular weight/ kD | 等电点 Point isoelectric(pI) | ||
|---|---|---|---|---|---|---|
| Soltu.DM.02G022850 | chr02 | 36 554 173-36 560 967 | 349 | 37.43 | 6.41 | |
| Soltu.DM.07G027060 | chr07 | 56 287 590-56 289 231 | 359 | 39.04 | 8.80 | |
| Soltu.DM.10G001180 | chr10 | 978 595-980 158 | 365 | 40.23 | 6.50 | |
| Soltu.DM.12G025770 | chr12 | 55 741 243-55 744 365 | 293 | 31.27 | 6.38 | |
表2 StSATs的染色体定位及理化性质
Table 2 Chromosomal localization and physicochemical properties of StSATs
| 基因ID Gene ID | 染色体定位 Chromosome localization | 氨基酸长度 Amino acid length | 相对分子量 Molecular weight/ kD | 等电点 Point isoelectric(pI) | ||
|---|---|---|---|---|---|---|
| Soltu.DM.02G022850 | chr02 | 36 554 173-36 560 967 | 349 | 37.43 | 6.41 | |
| Soltu.DM.07G027060 | chr07 | 56 287 590-56 289 231 | 359 | 39.04 | 8.80 | |
| Soltu.DM.10G001180 | chr10 | 978 595-980 158 | 365 | 40.23 | 6.50 | |
| Soltu.DM.12G025770 | chr12 | 55 741 243-55 744 365 | 293 | 31.27 | 6.38 | |
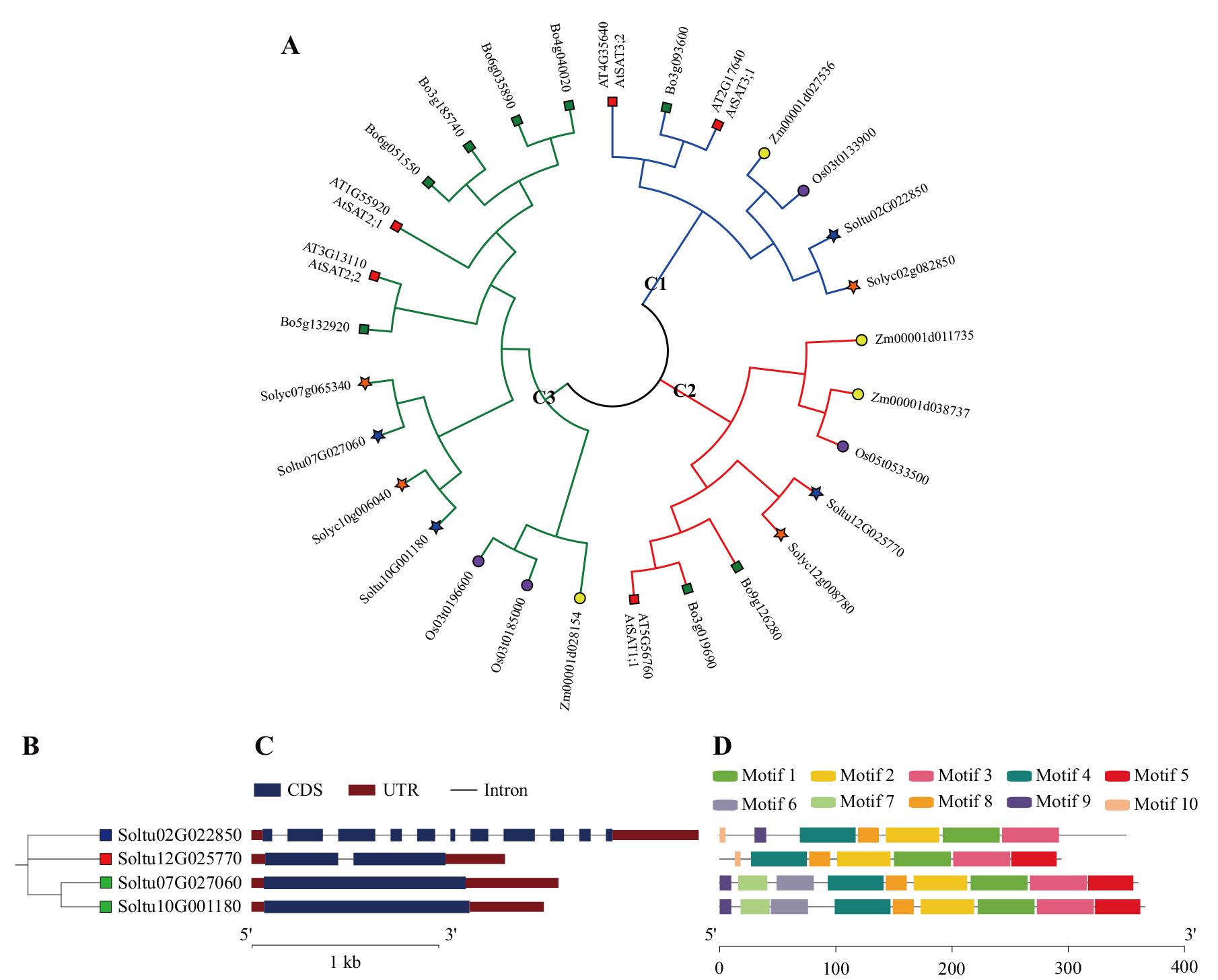
图1 多个物种中SAT蛋白的系统发育进化树以及StSAT基因家族成员的进化关系、基因结构和蛋白保守基序分析 A:多个物种中SAT蛋白的系统发育进化树,不同颜色的线代表3个亚族(蓝色代表C1亚族,红色代表C2亚族,绿色代表C3亚族),蓝色五角星表示马铃薯SAT家族成员(StSATs),橙色五角星表示番茄SAT家族成员(SlSATs),黄色圆圈表示玉米SAT家族成员ZmSATs,紫色圆圈表示水稻SAT家族成员(OsSATs),红色方块表示拟南芥SAT家族成员(AtSTAs),绿色方块表示甘蓝SAT家族成员(BoSATs);B:StSAT基因家族的进化树,蓝色方块代表C1亚族,红色方块代表C2亚族,绿色方块代表C3亚族;C:StSATs的基因结构,蓝色框表示外显子,黑线表示内含子,红色方框表示上/下游区域;D:StSATs蛋白保守基序
Fig. 1 Phylogenetic tree of SAT proteins in multiple species and analysis in evolutionary relationship, gene structure and protein conserved motifs of StSAT gene family members A: Phylogenetic tree of SAT proteins in multiple species. The different colored lines represent the three subfamilies(blue for C1, red for C2, and green for C3). The blue pentagrams indicate the potato SAT gene family member(StSATs), orange pentagrams indicate the tomato SAT gene family member(SlSATs), yellow circles indicate the corn SAT gene family member(ZmSATs), purple circles indicate the rice SAT gene family member(OsSATs), red squares indicate the Arabidopsis SAT gene family members(AtSTAs), and green squares indicate the cabbage SAT gene family member(BoSATs). B: Evolutionary tree of the StSAT gene family. The blue squares indicate the C1 subfamily, the red squares indicate the C2 subfamily, and the green squares indicate the C3 subfamily. C: Gene structure of StSATs. Blue boxes indicate exons, black lines indicate introns and red boxes indicate upstream/downstream regions. D: Protein conserved motif of StSATs
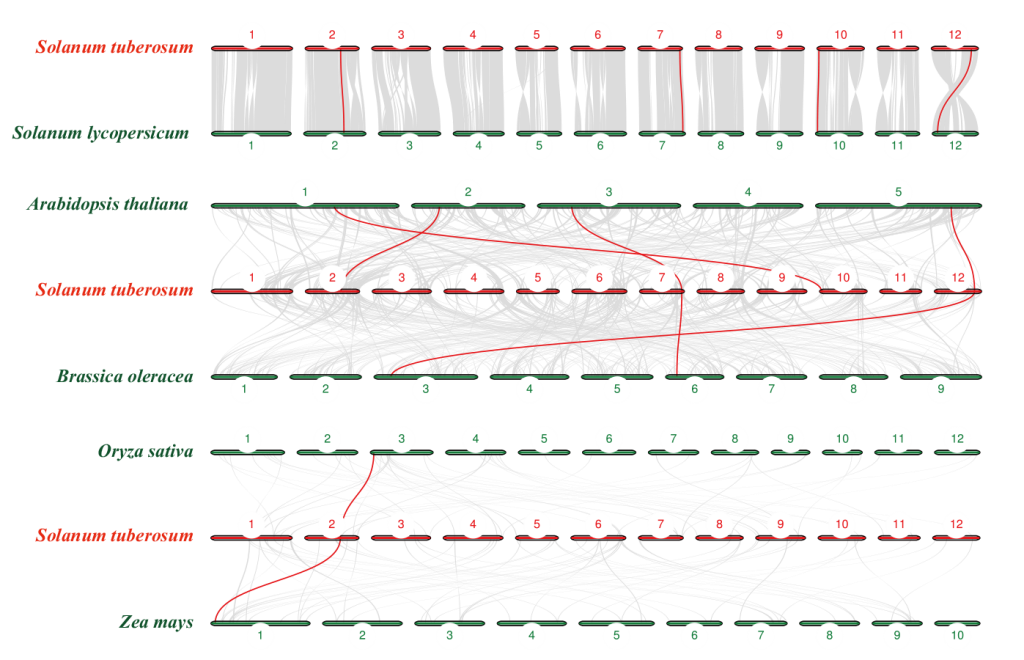
图2 多物种SAT基因的共线性分析 不同的数字编号代表不同物种的染色体,红线表示物种间的直系同源基因
Fig. 2 Collinearity analysis of SAT genes in multiple species Different numerical numbers indicate the chromosomes of different species, and the red lines indicate orthologous gene pairs between different species
| Gene ID1 | Gene ID2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Soltu02G022850 | Solyc02g082850 | 0.040 2 | 0.110 7 | 0.363 2 |
| Soltu12G025770 | Solyc12g008780 | 0.003 1 | 0.074 1 | 0.041 4 |
| Soltu07G027060 | Solyc07g065340 | 0.010 6 | 0.072 8 | 0.145 3 |
| Soltu10G001180 | Solyc10g006040 | 0.009 5 | 0.050 7 | 0.187 0 |
| Soltu02G022850 | AT2G17640 | 0.192 7 | 1.660 3 | 0.116 1 |
| Soltu12G025770 | AT5G56760 | 0.118 8 | 2.718 1 | 0.043 7 |
| Soltu07G027060 | AT3G13110 | 0.238 7 | 2.356 3 | 0.101 3 |
| Soltu10G001180 | AT1G55920 | 0.234 9 | 3.864 2 | 0.060 8 |
| Soltu12G025770 | Bo3g019690 | 0.122 7 | 3.050 0 | 0.040 2 |
| Soltu07G027060 | Bo6g035890 | 0.202 6 | 3.867 2 | 0.052 4 |
| Soltu02G022850 | Zm00001d027536 | 0.178 5 | 2.483 9 | 0.071 8 |
| Soltu02G022850 | Os03g04140 | 0.267 1 | 3.167 0 | 0.084 3 |
表3 马铃薯与其他物种间直系同源基因的Ka/Ks
Table 3 Ka/Ks ratios of orthologous genes between potato and other species
| Gene ID1 | Gene ID2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Soltu02G022850 | Solyc02g082850 | 0.040 2 | 0.110 7 | 0.363 2 |
| Soltu12G025770 | Solyc12g008780 | 0.003 1 | 0.074 1 | 0.041 4 |
| Soltu07G027060 | Solyc07g065340 | 0.010 6 | 0.072 8 | 0.145 3 |
| Soltu10G001180 | Solyc10g006040 | 0.009 5 | 0.050 7 | 0.187 0 |
| Soltu02G022850 | AT2G17640 | 0.192 7 | 1.660 3 | 0.116 1 |
| Soltu12G025770 | AT5G56760 | 0.118 8 | 2.718 1 | 0.043 7 |
| Soltu07G027060 | AT3G13110 | 0.238 7 | 2.356 3 | 0.101 3 |
| Soltu10G001180 | AT1G55920 | 0.234 9 | 3.864 2 | 0.060 8 |
| Soltu12G025770 | Bo3g019690 | 0.122 7 | 3.050 0 | 0.040 2 |
| Soltu07G027060 | Bo6g035890 | 0.202 6 | 3.867 2 | 0.052 4 |
| Soltu02G022850 | Zm00001d027536 | 0.178 5 | 2.483 9 | 0.071 8 |
| Soltu02G022850 | Os03g04140 | 0.267 1 | 3.167 0 | 0.084 3 |
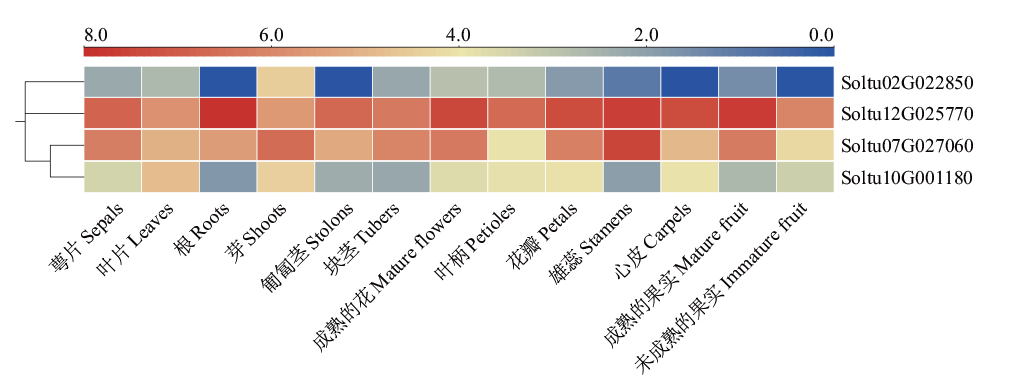
图4 StSATs在DM马铃薯不同组织部位中的表达 对4个StSATs在13个组织部位中的表达量取以2为底的对数,用log2FPKM绘制热图
Fig. 4 StSAT gene expressions in different tissues of DM potato The expressions of four StSATs in 13 tissue sites are logarithms base 2, and heat maps are plotted using log2FPKM
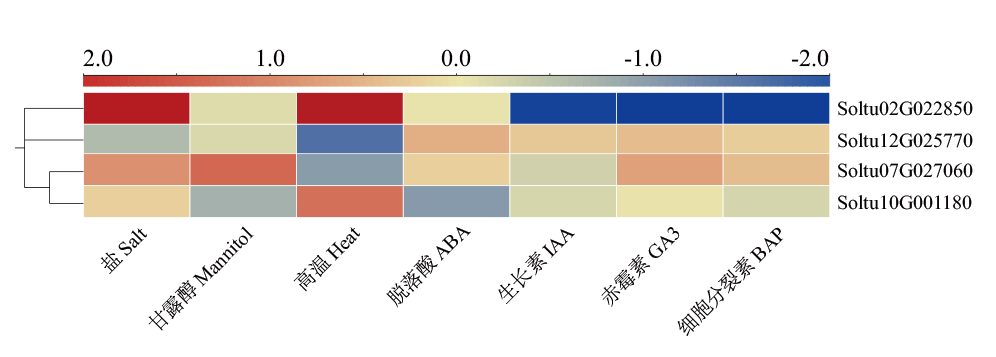
图5 StSATs在DM马铃薯非生物胁迫和激素处理下的表达 在非生物胁迫(盐、甘露醇和热胁迫)中使用处理与对照的比值,取以2为底的对数,用log2FC绘制热图
Fig. 5 StSAT gene expressions in DM potato under abiotic stress and hormone treatment The ratios of treatment to control are used for abiotic stresses(salt, mannitol, and heat stress), taken as base 2 logarithms and heat maps are plotted using log2FC
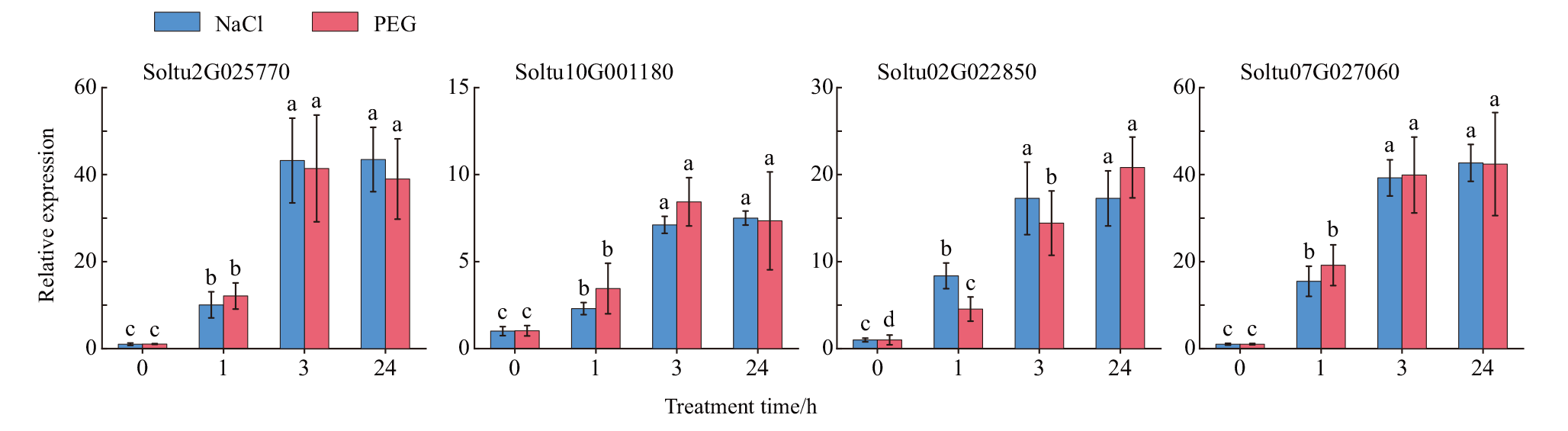
图6 StSAT基因在四倍体马铃薯NaCl和PEG处理下的相对表达水平 对4个马铃薯StSAT基因在NaCl和PEG处理下的表达进行了qPCR 分析。数据为3 个独立生物学重复的平均值(±SE)。柱状图上方的不同字母表示P<0.05 时的显著差异
Fig. 6 Relative expressions of StSAT genes under NaCl and PEG treatment in tetraploid potato The relative expressions of four StSAT genes under NaCl and PEG treatment was analyzed by qPCR. Data are means(±SEs)from three independent biological replicates. Different letters above the bar chart indicate significant differences at P<0.05
| [1] |
Beinert H. A tribute to sulfur[J]. Eur J Biochem, 2000, 267(18): 5657-5664.
pmid: 10971575 |
| [2] | Gerber J, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: components, mechanism and pathology[J]. Mitochondrion, 2002, 2(1/2): 71-86. |
| [3] |
Saito K. Sulfur assimilatory metabolism. The long and smelling road[J]. Plant Physiol, 2004, 136(1): 2443-2450.
doi: 10.1104/pp.104.046755 pmid: 15375200 |
| [4] |
Leustek T, Saito K. Sulfate transport and assimilation in plants[J]. Plant Physiol, 1999, 120(3): 637-644.
pmid: 10398698 |
| [5] | Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties[J]. J Plant Physiol, 2006, 163(3): 273-286. |
| [6] |
Romero LC, Ángeles Aroca M, Laureano-Marín AM, et al. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana[J]. Mol Plant, 2014, 7(2): 264-276.
doi: 10.1093/mp/sst168 pmid: 24285094 |
| [7] | Initiative AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana[J]. Nature, 2000, 408(6814): 796-815. |
| [8] |
Noji M, Inoue K, Kimura N, et al. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana[J]. J Biol Chem, 1998, 273(49): 32739-32745.
doi: 10.1074/jbc.273.49.32739 pmid: 9830017 |
| [9] |
Kawashima CG, Berkowitz O, Hell R, et al. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis[J]. Plant Physiol, 2005, 137(1): 220-230.
pmid: 15579666 |
| [10] |
Wirtz M, Berkowitz O, Droux M, et al. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction[J]. Eur J Biochem, 2001, 268(3): 686-693.
pmid: 11168407 |
| [11] | Kumar S, Kumar N, Alam N, et al. Crystal structure of serine acetyl transferase from Brucella abortus and its complex with coenzyme A[J]. Biochim Biophys Acta, 2014, 1844(10): 1741-1748. |
| [12] |
Francois JA, Kumaran S, Jez JM. Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex[J]. Plant Cell, 2006, 18(12): 3647-3655.
doi: 10.1105/tpc.106.047316 pmid: 17194764 |
| [13] | Jez JM, Dey S. The cysteine regulatory complex from plants and microbes: what was old is new again[J]. Curr Opin Struct Biol, 2013, 23(2): 302-310. |
| [14] |
Watanabe M, Mochida K, Kato T, et al. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis[J]. Plant Cell, 2008, 20(9): 2484-2496.
doi: 10.1105/tpc.108.060335 pmid: 18776059 |
| [15] |
Xiang XL, Wu YR, Planta J, et al. Overexpression of serine acetyltransferase in maize leaves increases seed-specific methionine-rich zeins[J]. Plant Biotechnol J, 2018, 16(5): 1057-1067.
doi: 10.1111/pbi.12851 pmid: 29044890 |
| [16] | Zhou H, Chen Y, Zhai FC, et al. Hydrogen sulfide promotes rice drought tolerance via reestablishing redox homeostasis and activation of ABA biosynthesis and signaling[J]. Plant Physiol Biochem, 2020, 155: 213-220. |
| [17] | Cao MJ, Wang Z, Zhao Q, et al. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana[J]. Plant J, 2014, 77(4): 604-615. |
| [18] |
Dominguez-Solis JR, He ZY, Lima A, et al. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts[J]. Proc Natl Acad Sci USA, 2008, 105(42): 16386-16391.
doi: 10.1073/pnas.0808204105 pmid: 18845687 |
| [19] |
Ahmad N, Malagoli M, Wirtz M, et al. Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots[J]. BMC Plant Biol, 2016, 16(1): 247.
pmid: 27829370 |
| [20] | Na G, Salt DE. Differential regulation of serine acetyltransferase is involved in nickel hyperaccumulation in Thlaspi goesingense[J]. J Biol Chem, 2011, 286(47): 40423-40432. |
| [21] | Liu DM, Li M, Guo T, et al. Functional characterization of the Serine acetyltransferase family genes uncovers the diversification and conservation of cysteine biosynthesis in tomato[J]. Front Plant Sci, 2022, 13: 913856. |
| [22] | Kurt F, Filiz E, Aydın A. Genome-wide identification of serine acetyltransferase(SAT)gene family in rice(Oryza sativa)and their expressions under salt stress[J]. Mol Biol Rep, 2021, 48(9): 6277-6290. |
| [23] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]//The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005: 571-607. |
| [24] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol, 2013, 30(5): 1229-1235.
doi: 10.1093/molbev/mst012 pmid: 23486614 |
| [25] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [26] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [27] | Chen CJ, Chen H, He YH, et al. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface[J]. bioRxiv, 2018. DOI: 10.1101/289660. |
| [28] | Liu Z, Liu YH, Coulter JA, et al. The WD40 gene family in potato(Solanum tuberosum L.): genome-wide analysis and identification of anthocyanin and drought-related WD40s[J]. Agronomy, 2020, 10(3): 401. |
| [29] | Li YM, Liang J, Zeng XZ, et al. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis[J]. Hortic Plant J, 2021, 7(2): 129-141. |
| [30] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana[J]. BMC Plant Biol, 2004, 4: 10. |
| [31] | Yeon JY, Yoo SJ, Takagi H, et al. A novel mitochondrial serine O-acetyltransferase, OpSAT1, plays a critical role in sulfur metabolism in the thermotolerant methylotrophic yeast Ogataea parapolymorpha[J]. Sci Rep, 2018, 8(1): 2377. |
| [32] | Venkatesh TV, Harrigan GG, Perez T, et al. Compositional assessments of key maize populations: B73 hybrids of the nested association mapping founder lines and diverse landrace inbred lines[J]. J Agric Food Chem, 2015, 63(21): 5282-5295. |
| [33] | Genisel M, Erdal S, Kizilkaya M. The mitigating effect of cysteine on growth inhibition in salt-stressed barley seeds is related to its own reducing capacity rather than its effects on antioxidant system[J]. Plant Growth Regul, 2015, 75(1): 187-197. |
| [34] | Sadak MS, Abd El-Hameid AR, Zaki FSA, et al. Physiological and biochemical responses of soybean(Glycine max L.) to cysteine application under sea salt stress[J]. Bull Natl Res Cent, 2019, 44(1): 1. |
| [35] | Zhang R, Xu C, Bao ZL, et al. Auxin alters sodium ion accumulation and nutrient accumulation by playing protective role in salinity challenged strawberry[J]. Plant Physiol Biochem, 2021, 164: 1-9. |
| [36] | Khalid A, Aftab F. Effect of exogenous application of IAA and GA3 on growth, protein content, and antioxidant enzymes of Solanum tuberosum L. grown in vitro under salt stress[J]. Vitro Cell Dev Biol Plant, 2020, 56(3): 377-389. |
| [1] | 王超, 白如仟, 管俊梅, 罗稷林, 何雪姣, 迟绍轶, 马玲. 马铃薯块茎变绿中StHY5对龙葵素合成的促进作用[J]. 生物技术通报, 2024, 40(9): 113-122. |
| [2] | 夏士轩, 耿泽栋, 祝光涛, 张春芝, 李大伟. 基于深度学习的马铃薯花粉活力快速检测[J]. 生物技术通报, 2024, 40(9): 123-130. |
| [3] | 毛向红, 卢瑶, 范向斌, 杜培兵, 白小东. 基于SSR荧光标记毛细管电泳的马铃薯品种遗传多样性分析及分子身份证构建[J]. 生物技术通报, 2024, 40(9): 131-140. |
| [4] | 袁兰, 黄娅楠, 张贝妮, 熊雨萌, 王洪洋. 基于流式细胞仪鉴定马铃薯倍性的高通量样品制备方法[J]. 生物技术通报, 2024, 40(9): 141-147. |
| [5] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [6] | 宋倩娜, 段永红, 冯瑞云. CRISPR/Cas9介导的高效四倍体马铃薯试管薯基因编辑体系的建立[J]. 生物技术通报, 2024, 40(9): 33-41. |
| [7] | 王柯然, 闫俊杰, 刘建凤, 高玉林. RNAi技术在马铃薯害虫防控中的应用和风险[J]. 生物技术通报, 2024, 40(9): 4-10. |
| [8] | 张小妹, 周南伶, 张赛行, 王超, 沈玉龙, 管俊梅, 马玲. 马铃薯StDREBs基因的克隆及其表达分析[J]. 生物技术通报, 2024, 40(9): 42-50. |
| [9] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [10] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [11] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| [12] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [13] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| [14] | 李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231. |
| [15] | 李勇慧, 鲍星星, 段一珂, 赵运霞, 于相丽, 陈尧, 张延召. 灵宝杜鹃bZIP家族全基因组鉴定及表达特征分析[J]. 生物技术通报, 2024, 40(8): 186-198. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||