








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 92-103.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0528
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
乔岩1,3( ), 杨芳1,2, 任盼荣1,2, 祁伟亮1,2, 安沛沛1,2, 李茜1,2, 李丹1,2, 肖俊飞4
), 杨芳1,2, 任盼荣1,2, 祁伟亮1,2, 安沛沛1,2, 李茜1,2, 李丹1,2, 肖俊飞4
收稿日期:2024-05-28
出版日期:2024-09-26
发布日期:2024-10-12
作者简介:乔 岩,男,博士,副教授,研究方向:马铃薯抗逆生理及分子生物学;E-mail: yanqiao@ldxy.edu.cn;乔岩同时为本文通信作者
基金资助:
QIAO Yan1,3( ), YANG Fang1,2, REN Pan-rong1,2, QI Wei-liang1,2, AN Pei-pei1,2, LI Qian1,2, LI Dan1,2, XIAO Jun-fei4
), YANG Fang1,2, REN Pan-rong1,2, QI Wei-liang1,2, AN Pei-pei1,2, LI Qian1,2, LI Dan1,2, XIAO Jun-fei4
Received:2024-05-28
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】1,4-二氢氧-2-石脑-CoA合成酶(1,4-dihydroxy-2-naphthoyl-CoA synthase,DHNS)基因是茄科植物糖苷生物碱合成代谢的潜在重要基因,开展马铃薯DHNS基因功能研究与验证,为低糖苷生物碱马铃薯品种(系)的选育提供基因和材料来源。【方法】利用RACE方法克隆得到马铃薯野生种恰柯薯(Solanum chacoense)ScDHNS基因,对其进行生物信息学分析和亚细胞定位,通过构建过表达载体pBWA(V)HS-DHNS转化马铃薯栽培种进行功能验证。【结果】ScDHNS cDNA序列开放阅读框1 023 bp,编码340个氨基酸,分子量为37.34 kD,等电点pI为8.592,具有典型的ECH保守结构域,属于烯酰水合酶超家族成员,在二穗短柄草(Brachypodium distachyon)、蒺藜苜蓿(Medicago truncatula)等植物基因组中都有其同源基因,且存在基因扩张和收缩事件。过表达ScDHNS基因后发现转化株ScDHNS和SGT1基因表达量显著上调,且表达量显著高于马铃薯WT植株。且对应转化植株的总糖苷生物碱含量显著高于马铃薯WT植株,最高可达到364.3 mg/kg,是对照的2.4倍。亚细胞定位结果显示ScDHNS定位于过氧化物酶体。【结论】马铃薯ScDHNS基因可能参与调控糖苷生物碱合成关键基因SGT1的表达,通过β-氧化途径和甲羟戊酸通路协同影响糖苷生物碱的合成,该基因与糖苷生物碱在亚细胞水平上的区室化有重要关系,对于培育低糖苷生物碱的马铃薯品种(系)具有重要的应用价值。
乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103.
QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species[J]. Biotechnology Bulletin, 2024, 40(9): 92-103.
| 引 物Primer | 序列Sequence(5'-3') |
|---|---|
| P1(g21236-5GSP1) | ACATTAAGGCGACCAAAACTTTCA |
| P2(g21236-5GSP2) | AACTTTCAAAATCAGCATAACCATCC |
| P3(outer primer) | GCTGTCAACGATACGCTACGTAAC |
| P4(inner primer) | GCTACGTAACGGCATGACAGTG |
| P5(g21236-3GSP1) | CCATTAGATAAGTTGGAGGCAG |
| P6(g21236-3GSP2) | GTCCTACAGCGATACGAGTGC |
| P7(outer primer) | TACCGTCGTTCCACTAGTGATTT |
| P8(inner primer) | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
表1 5'-和3'-RACE 所用引物
Table 1 Primers for the 5'- and 3'- RACE analysis
| 引 物Primer | 序列Sequence(5'-3') |
|---|---|
| P1(g21236-5GSP1) | ACATTAAGGCGACCAAAACTTTCA |
| P2(g21236-5GSP2) | AACTTTCAAAATCAGCATAACCATCC |
| P3(outer primer) | GCTGTCAACGATACGCTACGTAAC |
| P4(inner primer) | GCTACGTAACGGCATGACAGTG |
| P5(g21236-3GSP1) | CCATTAGATAAGTTGGAGGCAG |
| P6(g21236-3GSP2) | GTCCTACAGCGATACGAGTGC |
| P7(outer primer) | TACCGTCGTTCCACTAGTGATTT |
| P8(inner primer) | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 长度 Length/bp |
|---|---|---|---|
| SGT1-F | AAGCCACAATCCTCACTACCC | 57.7 | 132 |
| SGT1-R | AGGCAACCCAACTTCAGCAG | 59.2 | |
| ScDHNS-F | TTGATGATGGACATGCTGGACTTC | 58.0 | 84 |
| ScDHNS-R | TGCCTTCATTGCCTTCTTCAGTTC | 59.0 | |
| ef1-α-F | ATTCAAGTATGCCTGGGTGCT | 58.9 | 144 |
| ef1-α-R | TTCTTGATAAAGTCTCTGTGTCCG | 58.4 |
表2 荧光定量PCR引物序列
Table 2 Primers’ sequences for RT-qPCR
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 长度 Length/bp |
|---|---|---|---|
| SGT1-F | AAGCCACAATCCTCACTACCC | 57.7 | 132 |
| SGT1-R | AGGCAACCCAACTTCAGCAG | 59.2 | |
| ScDHNS-F | TTGATGATGGACATGCTGGACTTC | 58.0 | 84 |
| ScDHNS-R | TGCCTTCATTGCCTTCTTCAGTTC | 59.0 | |
| ef1-α-F | ATTCAAGTATGCCTGGGTGCT | 58.9 | 144 |
| ef1-α-R | TTCTTGATAAAGTCTCTGTGTCCG | 58.4 |
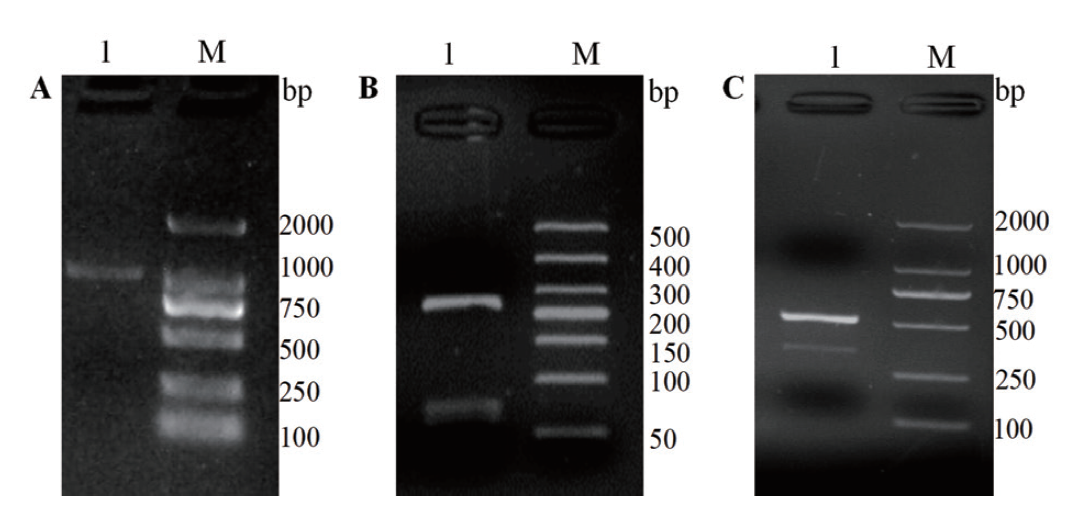
图1 ScDHNS 基因核心片段、5'-RACE 和3'-RACE 克隆 A:ScDHNS 基因核心片段克隆(泳道M:2000 DNA marker;泳道1:ScDHNS 基因CDS区域克隆片段);B:5'-RACE克隆(泳道M:500 DNA marker;泳道1:ScDHNS 基因5'-RACE克隆片段);C:3'-RACE克隆(泳道M:2000 DNA marker;泳道1:ScDHNS 基因3'-RACE克隆片段)
Fig. 1 The core fragment, 5'- and 3'- end fragments of gene ScDHNS A: The core fragment of ScDHNS gene(Lane M: 2000 DNA marker; lane 1: CDS cloning of ScDHNS gene). B: 5'-RACE end fragments of ScDHNS gene(Lane M: 500 DNA marker. Lane 1: 5'- RACE fragments of ScDHNS gene). C: 3'-RACE fragments of ScDHNS gene(Lane M: 2000 DNA marker. Lane 1: 5'- RACE fragments of ScDHNS gene)

图2 ScDHNS基因的核苷酸序列及其编码氨基酸序列 图中黑色方框代表着起始密码子和终止密码子,星号代表终止子
Fig. 2 Nucleotide sequences and deduced amino acid sequences of gene ScDHNS The black boxes indicate the initiation and termination codons, respectively. Asterisk indicates the stop codon

图3 ScDHNS与其他植物氨基酸序列的多重比较 直线表示ECH结构域,箭头为β折叠区,TT字母为β转角;螺旋线为α螺旋区和310-螺旋区(η)。各植物DHNS基因来源,恰柯薯(Solanum chacoense)ScDHNS、番茄(Solanum lycopersicum)SlDHNS、栽培马铃薯(Solanum tuberosum)StDHNS、渐狭叶烟草(Nicotiana attenuata)NaDHNS、二穗短柄草(Brachypodium distachyon)BdDHNS、拟南芥(Arabidopsis thaliana)AtDHNS、蒺藜苜蓿(Medicago truncatula)MtDHNS
Fig. 3 Multiple comparisons of amino acid sequences between ScDHNS and other plants The ECH domain is displayed as line. β-strands are rendered as arrows, strict β-turns as TT letters. The helix line indicates the α helix or 310- helix(η)area. DHNS gene sources: ScDHNS(Solanum chacoense), SlDHNS(Solanum lycopersicum), StDHNS(Solanum tuberosum), NaDHNS(Nicotiana attenuata), BdDHNS(Brachypodium distachyon), AtDHNS(Arabidopsis thaliana)and MtDHNS(Medicago truncatula)
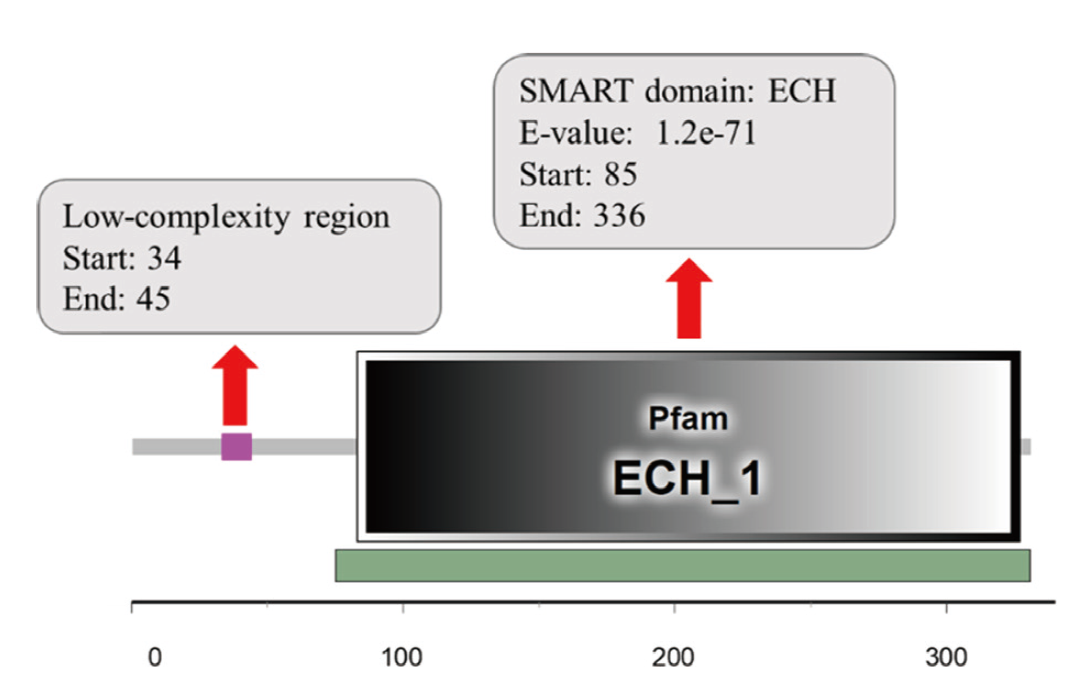
图4 ScDHNS蛋白质功能域分析 SMART domain 表示预测ScDHNS 蛋白结构域;刻度表示从起始密码子开始的密码子顺序
Fig. 4 Functional domain analysis of ScDHNS protein SMART domain indicates the predicted structural domain of ScDHNS. The scale indicates the order of the codon starting from the initial codon
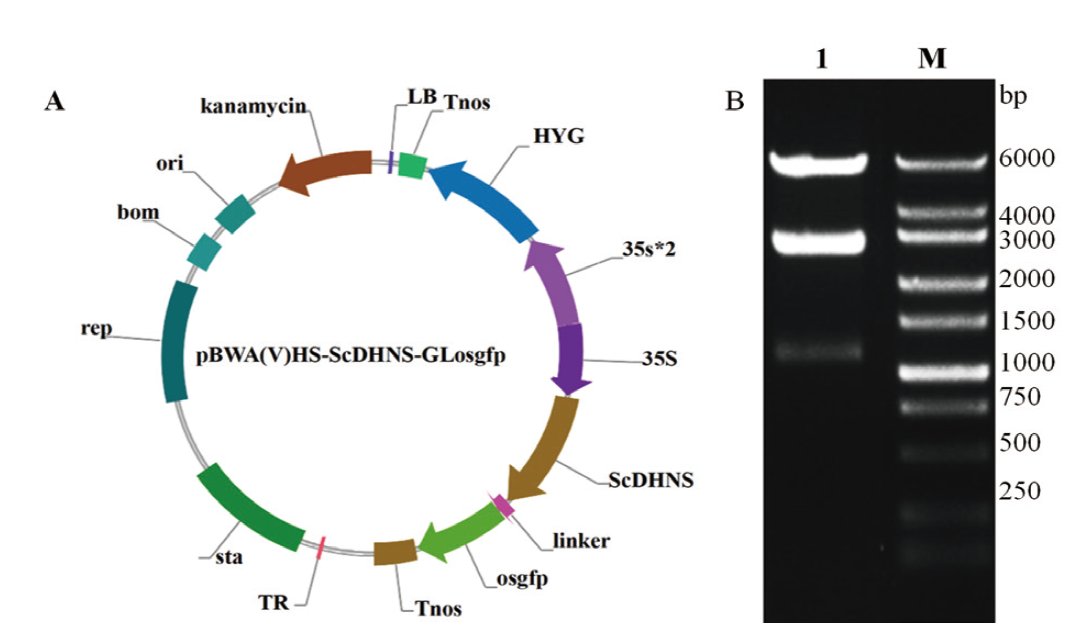
图6 pBWA-ScDHNS-Glosgfp亚细胞定位载体示意图及其酶切验证 A:pBWA-ScDHNS-Glosgfp亚细胞定位载体;B:pBWA-ScDHNS-Glosgfp载体酶切验证(M: DNA marker;泳道 1: 酶切后的载体)
Fig. 6 Profile of pBWA-ScDHNS-Glosgfp subcellular localization vector and its digestion verification A: Profile of pBWA-ScDHNS-Glosgfp subcellular localization vector. B: Digestion result of pBWA-ScDHNS-Glosgfp vector(Lane M: DNA marker. Lane 1: Digestion result of vector)

图7 ScDHNS在拟南芥原生质体中的亚细胞定位 A:拟南芥原生质体瞬时表达pBWA -Glosgfp空载体;B:拟南芥原生质体瞬时表达pBWA(V)HS-ScDHNS-Glosgfp重组载体
Fig. 7 Subcellular localization of ScDHNS in Arabidopsis protoplasts A: Empty vector pBWA-Glosgfp was transformed into Arabidopsis protoplasts. B: Recombinant vector pBWA(V)HS-ScDHNS-Glosgfp was transformed into Arabidopsis protoplasts
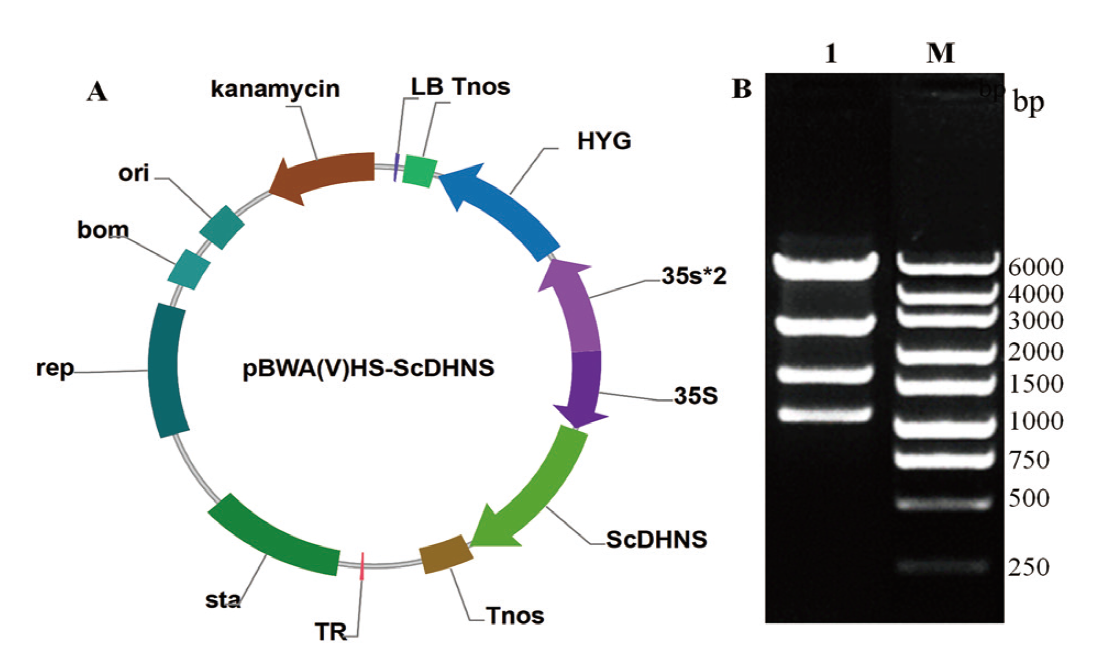
图8 pBWA(V)HS-ScDHNS表达载体及其酶切验证 A:pBWA(V)HS-ScDHNS表达载体;B:pBWA(V)HS-ScDHNS载体酶切验证(泳道M:DNA marker;泳道1:酶切后的载体)
Fig. 8 Profile of pBWA(V)HS-ScDHNS expressing vector and its enzymatic digestion verification A: pBWA(V)HS-ScDHNS express vector. B: Digestion result of pBWA(V)HS-ScDHNS vector(Lane M: DNA marker. Lane 1: Digestion result of vector)
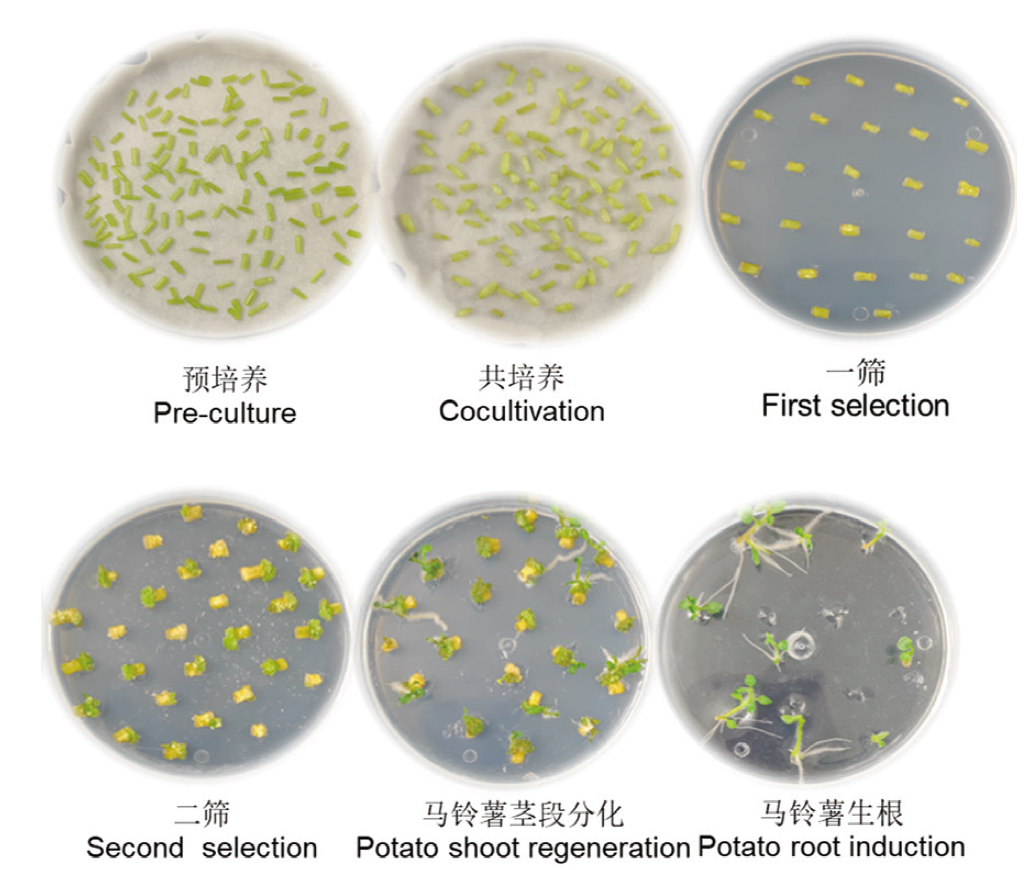
图9 农杆菌GV310(pBWA(V)HS-ScDHNS)转化马铃薯组培苗
Fig. 9 pBWA(V)HS-ScDHNS vector transformed into Agrobacterium tumefaciens GV3101 strain to tissue-cultivated seedlings of potato
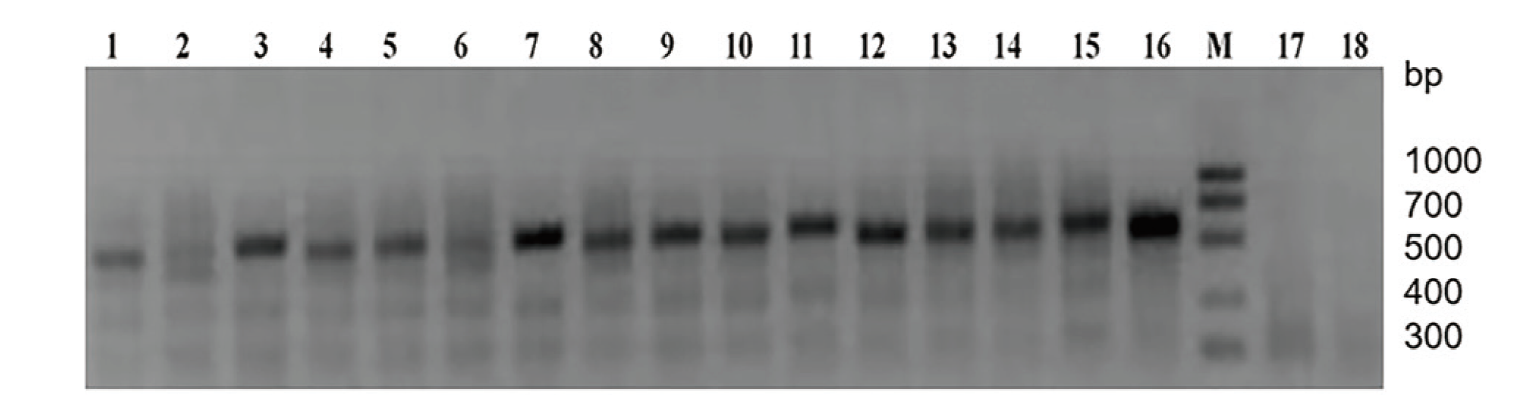
图10 pBWA(V)HS-ScDHNS转化马铃薯组培苗的PCR鉴定 M: DNA marker 1000;1-18:PCR产物
Fig. 10 PCR identification of potato tissue-cultivated seedlings transformed with pBWA(V)HS-ScDHNS M: DNA marker 1000. 1-18: PCR product
| [1] | 赵锋, 胡开明, 王晓斌, 等. 饲用马铃薯潜在产量的分析方法[J]. 草业科学, 2016, 33(11): 2326-2336. |
| Zhao F, Hu KM, Wang XB, et al. Analysis method of forage-use potato potential yields[J]. Pratacultural Sci, 2016, 33(11): 2326-2336. | |
| [2] | 夏善勇. 马铃薯渣与茎叶饲料化利用及研究进展[J]. 中国饲料, 2023(17): 137-143. |
| Xia SY. Research progress on forage utilization of potato residue and vine[J]. China Feed, 2023(17): 137-143. | |
| [3] | Cao QH, Dai WF, Li BC, et al. Sesquiterpenoids from the stems and leaves of Gochnatia decora[J]. Phytochem Lett, 2019, 30: 6-9. |
| [4] | 雒瑞瑞. 马铃薯茎叶和玉米秸/甜高粱混合青贮料的制作及其对瘤胃发酵特性的影响[D]. 兰州: 甘肃农业大学, 2018. |
| Luo RR. Preparation of mixed silage of potato stem and leaf and corn stalk/sweet sorghum and its influence on rumen fermentation characteristics[D]. Lanzhou: Gansu Agricultural University, 2018. | |
| [5] | Zhang SY, Deng JJ, Cui YF, et al. Effect of potato vine and leaf mixed silage to whole corn crops on rumen fermentation and the microbe of fatten Angus bulls[J]. Fermentation, 2023, 9(8): 704. |
| [6] | Qiao Y, Yang F, Li Q, et al. Combined small RNA and degradome sequencing reveals important roles of light-responsive microRNAs in wild potato(Solanum chacoense)[J]. Agronomy, 2023, 13(7): 1763. |
| [7] | Sakhare AV, Bess S, Mill LL. Isolation of solanine from potato leaves and evaluation of its antimicrobial activity[J]. Int J Sci Res, 2014, 3(11): 2052-2056. |
| [8] | Nützmann HW, Huang AC, Osbourn A. Plant metabolic clusters - from genetics to genomics[J]. New Phytol, 2016, 211(3): 771-789. |
| [9] | Zhao DK, Zhao Y, Chen SY, et al. Solanum steroidal glycoalkaloids: structural diversity, biological activities, and biosynthesis[J]. Nat Prod Rep, 2021, 38(8): 1423-1444. |
| [10] | Sonawane PD, Jozwiak A, Panda S, et al. ‘Hijacking’ core metabolism: a new panache for the evolution of steroidal glycoalkaloids structural diversity[J]. Curr Opin Plant Biol, 2020, 55: 118-128. |
| [11] |
Itkin M, Heinig U, Tzfadia O, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes[J]. Science, 2013, 341(6142): 175-179.
doi: 10.1126/science.1240230 pmid: 23788733 |
| [12] |
Cárdenas PD, Sonawane PD, Pollier J, et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway[J]. Nat Commun, 2016, 7: 10654.
doi: 10.1038/ncomms10654 pmid: 26876023 |
| [13] | McCue KF, Shepherd LVT, Allen PV, et al. Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase[J]. Plant Sci, 2005, 168(1): 267-273. |
| [14] | Widhalm JR, Ducluzeau AL, Buller NE, et al. Phylloquinone(vitamin K(1))biosynthesis in plants: two peroxisomal thioesterases of Lactobacillales origin hydrolyze 1, 4-dihydroxy-2-naphthoyl-CoA[J]. Plant J, 2012, 71(2): 205-215. |
| [15] | Kumar S, Tripathi J, Srivastava AK, et al. Molecular mechanism of antimutagenicity by an ethoxy-substituted phylloquinone(vitamin K1 derivative)from spinach(Spinacea oleracea L.)[J]. Chem Biol Interact, 2020, 330: 109216. |
| [16] |
Mweetwa AM, Hunter D, Poe R, et al. Steroidal glycoalkaloids in Solanum chacoense[J]. Phytochemistry, 2012, 75: 32-40.
doi: 10.1016/j.phytochem.2011.12.003 pmid: 22217745 |
| [17] |
Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends[J]. Anal Biochem, 1995, 227(2): 255-273.
pmid: 7573945 |
| [18] | Beaujean A, Sangwan RS, Lecardonnel A, et al. Agrobacterium -mediated transformation of three economically important potato cultivars using sliced internodal explants: an efficient protocol of transformation[J]. J Exp Bot, 1998, 49(326): 1589-1595. |
| [19] | Grunenfelder LA, Knowles LO, Hiller LK, et al. Glycoalkaloid development during greening of fresh market potatoes(Solanum tuberosum L.)[J]. J Agric Food Chem, 2006, 54(16): 5847-5854. |
| [20] | Bolser DM, Kerhornou A, Walts B, et al. Triticeae resources in Ensembl Plants[J]. Plant Cell Physiol, 2015, 56(1): e3. |
| [21] |
De Bie T, Cristianini N, Demuth JP, et al. CAFE: a computational tool for the study of gene family evolution[J]. Bioinformatics, 2006, 22(10): 1269-1271.
doi: 10.1093/bioinformatics/btl097 pmid: 16543274 |
| [22] | Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms[J]. Nat Protoc, 2008, 3(2): 153-162. |
| [23] | 乔岩. 马铃薯光诱导糖苷生物碱代谢相关miRNAs的鉴定与功能分析[D]. 兰州: 甘肃农业大学, 2017. |
| Qiao Y. Identification and functional analysis of miRNAs related to photoinduced glycoside alkaloid metabolism in potato[D]. Lanzhou: Gansu Agricultural University, 2017. | |
| [24] | Huang J, Xu WB, Zhai JW, et al. Nuclear phylogeny and insights into whole-genome duplications and reproductive development of Solanaceae plants[J]. Plant Commun, 2023, 4(4): 100595. |
| [25] | Dalwani S, Wierenga RK. Enzymes of the crotonase superfamily: diverse assembly and diverse function[J]. Curr Opin Struct Biol, 2023, 82: 102671. |
| [26] | Greenhagen BT, Schoenbeck MA, Yeo YS, et al. Chapter ten The chemical wizardry of isoprenoid metabolism in plants[M]// Recent Advances in Phytochemistry. Amsterdam: Elsevier, 2003: 231-251. |
| [27] | del Río LA, Schrader M. Proteomics of peroxisomes: identifying novel functions and regulatory networks[M]. Singapore: Springer Singapore, 2018:3-45. |
| [28] |
Pan RH, Liu J, Wang SS, et al. Peroxisomes: versatile organelles with diverse roles in plants[J]. New Phytol, 2020, 225(4): 1410-1427.
doi: 10.1111/nph.16134 pmid: 31442305 |
| [29] |
Li HB, Brouwer M, Pup ED, et al. Allelic variation in the autotetraploid potato: genes involved in starch and steroidal glycoalkaloid metabolism as a case study[J]. BMC Genomics, 2024, 25(1): 274.
doi: 10.1186/s12864-024-10186-5 pmid: 38475714 |
| [1] | 邢丽南, 张艳芳, 葛明然, 赵令敏, 陈妍, 霍秀文. 山药DoWRKY40基因表达特征分析及互作蛋白筛选[J]. 生物技术通报, 2024, 40(8): 118-128. |
| [2] | 林彤, 袁程, 董陈文华, 曾孟琼, 杨燕, 毛自朝, 林春. 藜麦配子发育相关基因CqSTK的筛选及功能分析[J]. 生物技术通报, 2024, 40(8): 83-94. |
| [3] | 沈真辉, 曹瑶, 杨林雷, 罗祥英, 子灵山, 陆青青, 李荣春. 金耳和毛韧革菌麦角硫因生物合成基因的克隆及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 259-272. |
| [4] | 黄丹, 姜山, 彭涛. 褐角苔FfCYP98基因克隆及其功能分析[J]. 生物技术通报, 2024, 40(7): 273-284. |
| [5] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [6] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [7] | 李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202. |
| [8] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [9] | 闫欢欢, 尚怡彤, 王丽红, 田学琴, 廖海艳, 曾斌, 胡志宏. 米曲霉异源表达合成虫草素[J]. 生物技术通报, 2024, 40(6): 290-298. |
| [10] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [11] | 潘萍萍, 徐志浩, 张怡雯, 李青, 王忠华. 多花黄精查尔酮合酶PcCHS的原核表达、亚细胞定位及表达分析[J]. 生物技术通报, 2024, 40(5): 280-289. |
| [12] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [13] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [14] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [15] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||