








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 320-328.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0159
收稿日期:2024-02-18
出版日期:2024-08-26
发布日期:2024-09-05
通讯作者:
罗仍卓么,女,博士,副教授,研究方向:奶牛基因表达调控与分子育种;E-mail: luorenzhuoma@nxu.edu.cn作者简介:周冉,女,硕士研究生,研究方向:动物遗传育种;E-mail: zhouran18243320047@163.com
基金资助:
ZHOU Ran( ), WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma(
), WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma( )
)
Received:2024-02-18
Published:2024-08-26
Online:2024-09-05
摘要:
【目的】长链非编码RNA(lncRNA)可参与机体炎症反应,但其在奶牛乳房炎中的表达模式及分子调控机制仍不完全清楚。分析金黄色葡萄球菌(Staphylococcus aureus, S.aureus)型乳房炎奶牛乳腺组织中的差异表达lncRNA(DElncRNA)及其功能,为后续深入研究提供支撑。【方法】利用转录组测序(RNA-Seq)技术和生物信息学方法对健康奶牛和S. aureus诱导乳房炎的奶牛乳腺组织进行lncRNA测序、DElncRNA分析及GO和KEGG功能富集分析。【结果】在两组乳腺组织中共检测到1 012个lncRNA。相比于健康奶牛组,S. aureus诱导组筛选到75个表达上调的和114个表达下调的lncRNA。GO和KEGG富集分析结果表明,上述DElncRNA可通过免疫相关信号通路而调节奶牛乳房炎症。进一步分析发现,DElncRNA TCONS_00042123、TCONS_00068055、TCONS_00108420和TCONS_00076361可能参与MAPK、NLR、Jak-STAT和TLR信号通路,进而调节S. aureus型奶牛乳房炎的发生与发展。【结论】在S. aureus诱导炎症的奶牛乳腺组织中获得了189个DElncRNA,可能通过潜在靶基因调控S. aureus型奶牛乳房炎的发生与发展过程。
周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328.
ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis[J]. Biotechnology Bulletin, 2024, 40(8): 320-328.
| 长链非编码RNA lncRNA | 引物序列 Primer sequence(5'-3') | 产物长度 Product length/bp |
|---|---|---|
| TCONS_00018117 | F: TGCCAGGACAAGGAGAAA R: TCAGCACCTAAACCACAAGA | 100 |
| TCONS_00096253 | F: GGAGCGTAAGTAGGAAGCG R: AATGGTCCTGAATAGGGGTT | 92 |
| TCONS_00013236 | F: ATGGAAAAACGGACTCGG R: CGGTCTGCCTGAGTCTTG | 88 |
| TCONS_00026262 | F: AATGCCAGGGCTCCAGTT R: CTCAGATGGACAGTATTCCCTTT | 89 |
| TCONS_00018115 | F: GCCAGGACAAGGAGAAACA R: TCAGCACCTAAACCACAAGAC | 100 |
| TCONS_00113348 | F: GGTTCTATTGGATACTGGACAT R: CCTTGCTCTGCTCTTCTTTA | 100 |
| TCONS_00120135 | F: GCCTTTCATTTTCAAGAGCAT R: TGTCAAGGGAAGGGTGTCTG | 90 |
| TCONS_00044738 | F: AGGGCTGTTTACCTGCTTA R: GGTGATTTCGGTTCCTCTA | 93 |
| TCONS_00032949 | F: CAAGACGGCGACTTAGAAA R: ATGCCATCAAGTGGAACAA | 94 |
| TCONS_00124429 | F: GTCCACCTCCACCCTACA R: ATGCTCACCGAAGTCAAAG | 100 |
| TCONS_00117470 | F: TGTGCTGGACATAAATCGTG R: AGGTTCTGGGATTGTCTGC | 93 |
| TCONS_00088875 | F: GATGGTGCTGCTTCAGGATG R: TGGAGGCTACGCCAGGTT | 94 |
| GAPDH a | F: GGCATCGTGGAGGGACTTATG R: CCAGTGAGCTTCCCGTTGAG | 185 |
| RPS18 b | F: GTGGTGTTGAGGAAAGCAGACA R: TGATCACACGTTCCACCTCATC | 79 |
表1 用于lncRNA表达量检测的RT-qPCR引物
Table 1 RT-qPCR primers for lncRNA expression detection
| 长链非编码RNA lncRNA | 引物序列 Primer sequence(5'-3') | 产物长度 Product length/bp |
|---|---|---|
| TCONS_00018117 | F: TGCCAGGACAAGGAGAAA R: TCAGCACCTAAACCACAAGA | 100 |
| TCONS_00096253 | F: GGAGCGTAAGTAGGAAGCG R: AATGGTCCTGAATAGGGGTT | 92 |
| TCONS_00013236 | F: ATGGAAAAACGGACTCGG R: CGGTCTGCCTGAGTCTTG | 88 |
| TCONS_00026262 | F: AATGCCAGGGCTCCAGTT R: CTCAGATGGACAGTATTCCCTTT | 89 |
| TCONS_00018115 | F: GCCAGGACAAGGAGAAACA R: TCAGCACCTAAACCACAAGAC | 100 |
| TCONS_00113348 | F: GGTTCTATTGGATACTGGACAT R: CCTTGCTCTGCTCTTCTTTA | 100 |
| TCONS_00120135 | F: GCCTTTCATTTTCAAGAGCAT R: TGTCAAGGGAAGGGTGTCTG | 90 |
| TCONS_00044738 | F: AGGGCTGTTTACCTGCTTA R: GGTGATTTCGGTTCCTCTA | 93 |
| TCONS_00032949 | F: CAAGACGGCGACTTAGAAA R: ATGCCATCAAGTGGAACAA | 94 |
| TCONS_00124429 | F: GTCCACCTCCACCCTACA R: ATGCTCACCGAAGTCAAAG | 100 |
| TCONS_00117470 | F: TGTGCTGGACATAAATCGTG R: AGGTTCTGGGATTGTCTGC | 93 |
| TCONS_00088875 | F: GATGGTGCTGCTTCAGGATG R: TGGAGGCTACGCCAGGTT | 94 |
| GAPDH a | F: GGCATCGTGGAGGGACTTATG R: CCAGTGAGCTTCCCGTTGAG | 185 |
| RPS18 b | F: GTGGTGTTGAGGAAAGCAGACA R: TGATCACACGTTCCACCTCATC | 79 |
| 样品 Sample | 质控后的读段数 Clean reads | 对比到参考基因组的读段数 Mapped reads | 参考基因组上有唯一比对位置的读段数 Uniq mapped reads | 参考基因组上有多个比对位置的读段数 Multiple mapped reads | GC /% | Q30 /% |
|---|---|---|---|---|---|---|
| M.C | 128 303 002 | 108 631 698 (84.67%) | 97 000 181 (89.29%) | 11 631 517 (10.71%) | 52.30 | 85.60 |
| M.S | 108 776 484 | 91 300 177 (83.93%) | 84 635 626 (92.70%) | 6 664 551 (7.30%) | 50.84 | 85.16 |
表2 测序文库质量统计
Table 2 Quality statistics of sequencing libraries
| 样品 Sample | 质控后的读段数 Clean reads | 对比到参考基因组的读段数 Mapped reads | 参考基因组上有唯一比对位置的读段数 Uniq mapped reads | 参考基因组上有多个比对位置的读段数 Multiple mapped reads | GC /% | Q30 /% |
|---|---|---|---|---|---|---|
| M.C | 128 303 002 | 108 631 698 (84.67%) | 97 000 181 (89.29%) | 11 631 517 (10.71%) | 52.30 | 85.60 |
| M.S | 108 776 484 | 91 300 177 (83.93%) | 84 635 626 (92.70%) | 6 664 551 (7.30%) | 50.84 | 85.16 |
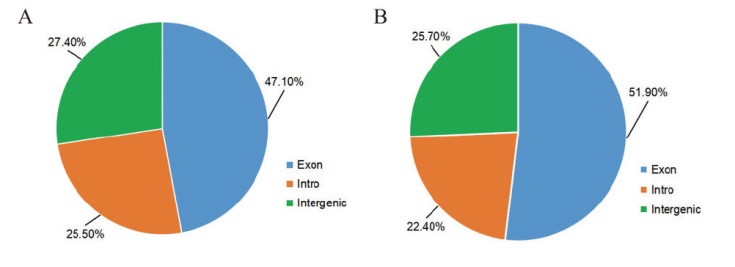
图1 Mapped reads在基因组中的分布 A:对照组中Mapped reads分布;B:S. aureus诱导组中Mapped reads分布
Fig. 1 Distribution of mapped reads in the genome A: Distribution of mapped reads in the control group; B: distribution of mapped reads in the S. aureus-induced group
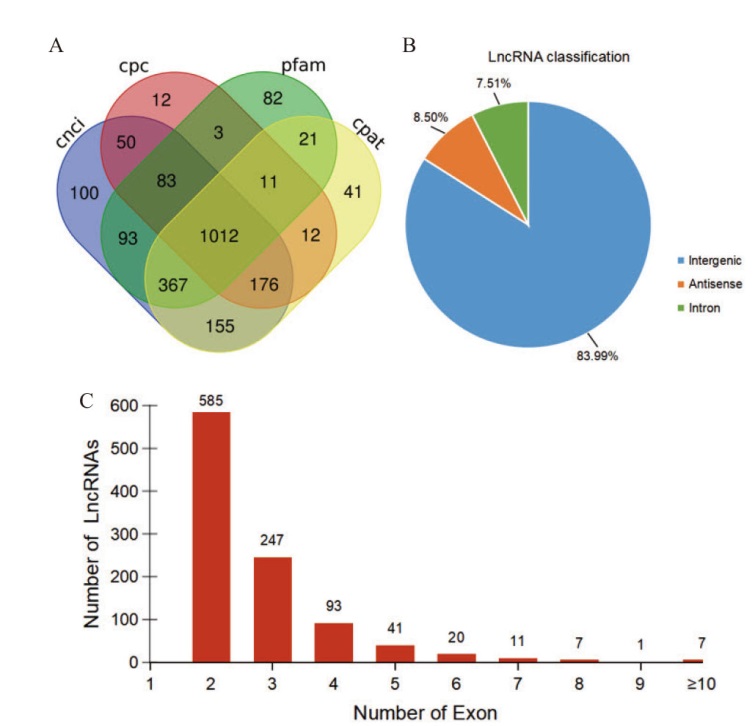
图2 lncRNA的鉴定 A: lncRNA预测韦恩图;B: lncRNA分类;C: lncRNA外显子数量分布
Fig. 2 LncRNA identification A: Venn diagram of lncRNA prediction; B: classification of lncRNA; C: distribution of lncRNA exon number
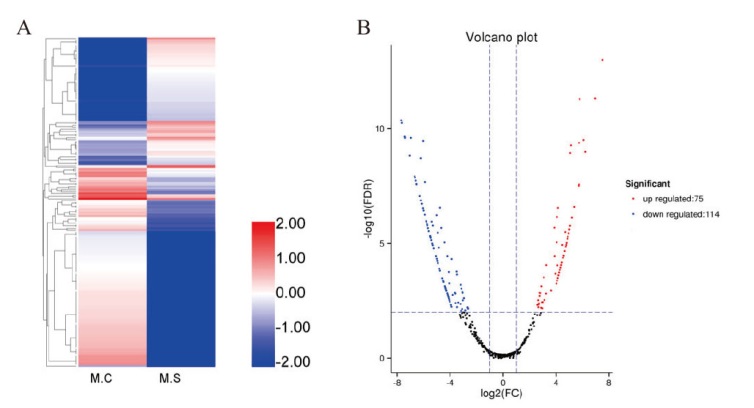
图3 lncRNA的差异表达分析 A:DElncRNA的聚类热图;B:DElncRNA的火山图
Fig. 3 Differential expression analysis of lncRNA A: Clustering heatmap of DElncRNA; B: volcano plot of DElncRNA
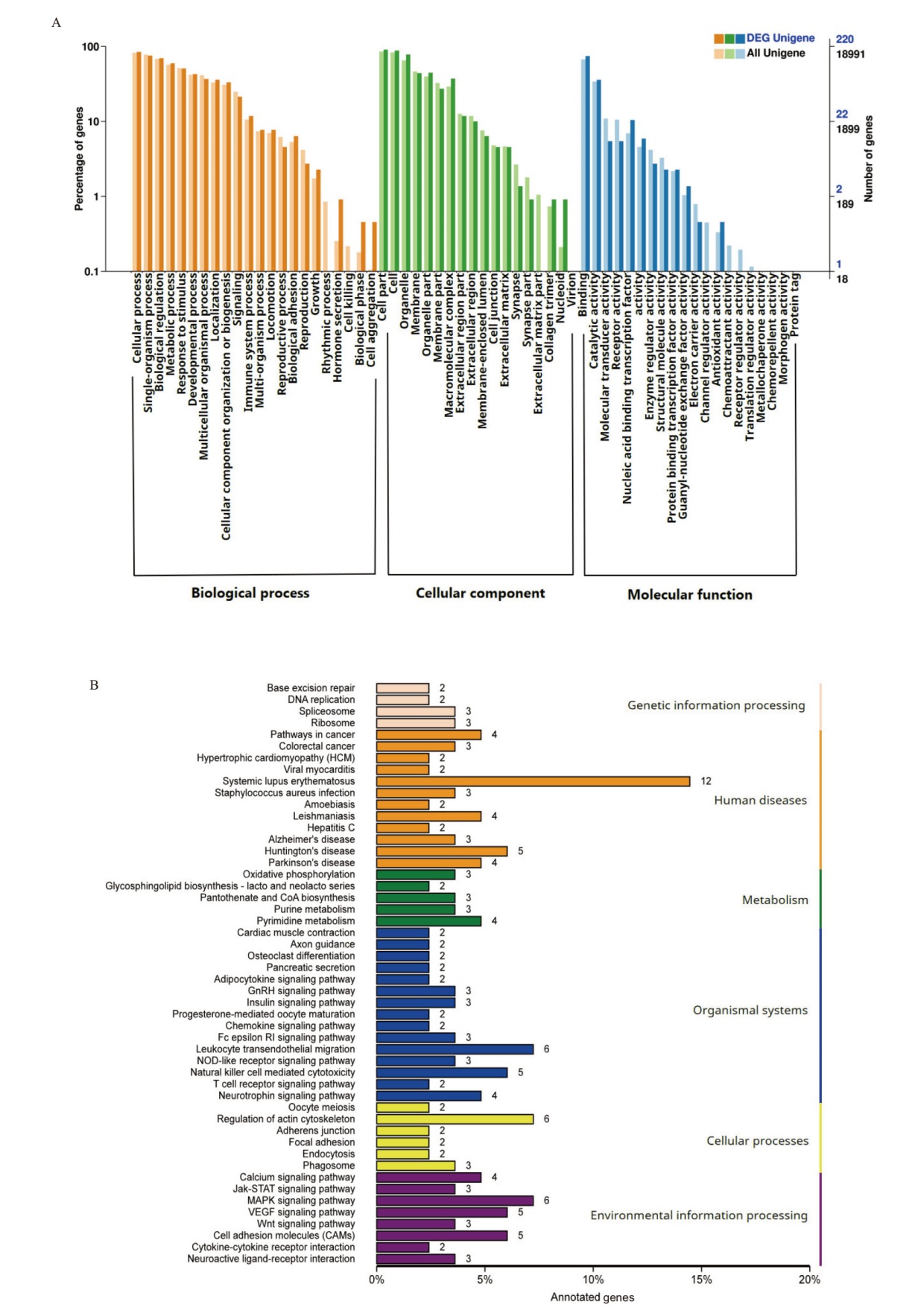
图5 DElncRNA靶基因的GO和KEGG富集分析 A:DElncRNA靶基因的GO注释分析;B:DElncRNA靶基因的KEGG富集分析
Fig. 5 GO annotation and KEGG enrichment analysis of DElncRNA target genes A: GO annotation analysis of DElncRNA target genes; B: KEGG enrichment analysis of DElncRNA target genes
| [1] |
Doehring C, Sundrum A. The informative value of an overview on antibiotic consumption, treatment efficacy and cost of clinical mastitis at farm level[J]. Prev Vet Med, 2019, 165: 63-70.
doi: S0167-5877(17)30827-9 pmid: 30851929 |
| [2] |
Royster E, Wagner S. Treatment of mastitis in cattle[J]. Vet Clin North Am Food Anim Pract, 2015, 31(1): 17-46.
doi: 10.1016/j.cvfa.2014.11.010 |
| [3] |
Ruegg PL. A 100-Year Review: Mastitis detection, management, and prevention[J]. J Dairy Sci, 2017, 100(12): 10381-10397.
doi: S0022-0302(17)31032-9 pmid: 29153171 |
| [4] | Gorji AE, Roudbari Z, Sadeghi B, et al. Transcriptomic analysis on the promoter regions discover gene networks involving mastitis in cattle[J]. Microb Pathog, 2019, 137: 103801. |
| [5] | Chen Y, Yang J, Huang Z, et al. Vitexin mitigates Staphylococcus aureus-induced mastitis via regulation of ROS/ER stress/NF- κ B/MAPK pathway[J]. Oxid Med Cell Longev, 2022, 2022: 7977433. |
| [6] |
Wu YH, He T, Fu YH, et al. Corynoline protects lipopolysaccharide-induced mastitis through regulating AKT/GSK3β/Nrf2 signaling pathway[J]. Environ Toxicol, 2021, 36(12): 2493-2499.
doi: 10.1002/tox.23362 pmid: 34477289 |
| [7] | Ran X, Yan Z, Yang YX, et al. Dioscin improves pyroptosis in LPS-induced mice mastitis by activating AMPK/Nrf2 and inhibiting the NF- κB signaling pathway[J]. Oxid Med Cell Longev, 2020, 2020: 8845521. |
| [8] | Mishra SK, Zhong ZM, Wang H. Hundreds of LncRNAs display circadian rhythmicity in zebrafish larvae[J]. Cells, 2021, 10(11): 3173. |
| [9] |
Tang H, Yuan S, Chen T, et al. Development of an immune-related lncRNA-miRNA-mRNA network based on competing endogenous RNA in periodontitis[J]. J Clin Periodontol, 2021, 48(11): 1470-1479.
doi: 10.1111/jcpe.13537 pmid: 34409632 |
| [10] | Cao HL, Liu ZJ, Huang PL, et al. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206[J]. Eur Rev Med Pharmacol Sci, 2019, 23(3): 1012-1021. |
| [11] |
Tian F, Wang JH, Zhang ZH, et al. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis[J]. Biol Res, 2020, 53(1): 9.
doi: 10.1186/s40659-020-00275-6 pmid: 32066502 |
| [12] | Lin CJ, Zhu YF, Hao ZY, et al. Genome-wide analysis of LncRNA in bovine mammary epithelial cell injuries induced by Escherichia coli and Staphylococcus aureus[J]. Int J Mol Sci, 2021, 22(18): 9719. |
| [13] | Chen Y, Jing HY, Chen MY, et al. Transcriptional profiling of exosomes derived from Staphylococcus aureus-infected bovine mammary epithelial cell line MAC-T by RNA-seq analysis[J]. Oxid Med Cell Longev, 2021, 2021: 8460355. |
| [14] |
Tong C, Chen QL, Zhao LL, et al. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands[J]. BMC Genomics, 2017, 18(1): 468.
doi: 10.1186/s12864-017-3858-4 pmid: 28629368 |
| [15] |
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2[J]. Nat Methods, 2012, 9(4): 357-359.
doi: 10.1038/nmeth.1923 pmid: 22388286 |
| [16] | Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions[J]. Genome Biol, 2013, 14(4): R36. |
| [17] |
Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks[J]. Nat Protoc, 2012, 7(3): 562-578.
doi: 10.1038/nprot.2012.016 pmid: 22383036 |
| [18] |
Bougarn S, Cunha P, Gilbert FB, et al. Technical note: validation of candidate reference genes for normalization of quantitative PCR in bovine mammary epithelial cells responding to inflammatory stimuli[J]. J Dairy Sci, 2011, 94(5): 2425-2430.
doi: 10.3168/jds.2010-3859 pmid: 21524534 |
| [19] |
Tripurani SK, Xiao CD, Salem M, et al. Cloning and analysis of fetal ovary microRNAs in cattle[J]. Anim Reprod Sci, 2010, 120(1-4): 16-22.
doi: 10.1016/j.anireprosci.2010.03.001 pmid: 20347535 |
| [20] |
Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs[J]. Nat Rev Genet, 2020, 21(2): 102-117.
doi: 10.1038/s41576-019-0184-5 pmid: 31729473 |
| [21] |
Li JW, Ma W, Zeng P, et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs[J]. Brief Bioinform, 2015, 16(5): 806-812.
doi: 10.1093/bib/bbu048 pmid: 25524864 |
| [22] | Mao XZ, Cai T, Olyarchuk JG, et al. Automated genome annotation and pathway identification using the KEGG Orthology(KO)as a controlled vocabulary[J]. Bioinformatics, 2005, 21(19): 3787-3793. |
| [23] | Sun YJ, Zhao TQ, Ma YY, et al. Multiple roles of LncRNA-BMNCR on cell proliferation and apoptosis by targeting miR-145/CBFB axis in BMECs[J]. Vet Q, 2023, 43(1): 1-11. |
| [24] |
Zhou M, Barkema HW, Gao J, et al. MicroRNA miR-223 modulates NLRP3 and Keap1, mitigating lipopolysaccharide-induced inflammation and oxidative stress in bovine mammary epithelial cells and murine mammary glands[J]. Vet Res, 2023, 54(1): 78.
doi: 10.1186/s13567-023-01206-5 pmid: 37710276 |
| [25] | Chen Z, Liang Y, Lu QY, et al. Cadmium promotes apoptosis and inflammation via the circ08409/miR-133a/TGFB2 axis in bovine mammary epithelial cells and mouse mammary gland[J]. Ecotoxicol Environ Saf, 2021, 222: 112477. |
| [26] | Chen Y, Yang J, Huang Z, et al. Exosomal lnc-AFTR as a novel translation regulator of FAS ameliorates Staphylococcus aureus-induced mastitis[J]. BioFactors, 2022, 48(1): 148-163. |
| [27] | Feng F, Li YX, Wang JP, et al. LncRNA CA12-AS1 targets miR-133a to promote LPS-induced inflammatory response in bovine mammary epithelial cells[J]. Int J Biol Macromol, 2024, 261(Pt 1): 129710. |
| [28] | Bai ZX, Wu YZ, Cai WD, et al. High-throughput analysis of lncRNA in cows with naturally infected Staphylococcus aureus mammary gland[J]. Anim Biotechnol, 2023, 34(7): 2166-2174. |
| [29] | Wang JP, Hu QC, Yang J, et al. Differential expression profiles of lncRNA following LPS-induced inflammation in bovine mammary epithelial cells[J]. Front Vet Sci, 2021, 8: 758488. |
| [30] | Jing HY, Chen Y, Qiu CW, et al. LncRNAs transcriptome analysis revealed potential mechanisms of selenium to mastitis in dairy cows[J]. Biol Trace Elem Res, 2022, 200(10): 4316-4324. |
| [31] | Zhang YH, Xu YQ, Chen BW, et al. Selenium deficiency promotes oxidative stress-induced mastitis via activating the NF-κB and MAPK pathways in dairy cow[J]. Biol Trace Elem Res, 2022, 200(6): 2716-2726. |
| [32] |
Li JD, Yin P, Gong P, et al. 8-Methoxypsoralen protects bovine mammary epithelial cells against lipopolysaccharide-induced inflammatory injury via suppressing JAK/STAT and NF-κB pathway[J]. Microbiol Immunol, 2019, 63(10): 427-437.
doi: 10.1111/1348-0421.12730 pmid: 31313848 |
| [33] |
Saxena M, Yeretssian G. NOD-like receptors: master regulators of inflammation and cancer[J]. Front Immunol, 2014, 5: 327.
doi: 10.3389/fimmu.2014.00327 pmid: 25071785 |
| [34] | Kiewiet MBG, Dekkers R, Gros M, et al. Toll-like receptor mediated activation is possibly involved in immunoregulating properties of cow's milk hydrolysates[J]. PLoS One, 2017, 12(6): e0178191. |
| [35] | Xu P, Xu XB, Fotina H, et al. Anti-inflammatory effects of chlorogenic acid from Taraxacum officinale on LTA-stimulated bovine mammary epithelial cells via the TLR2/NF-κB pathway[J]. PLoS One, 2023, 18(3): e0282343. |
| [36] | Cao FQ, Zhou W, Liu GH, et al. Staphylococcus aureus peptidoglycan promotes osteoclastogenesis via TLR2-mediated activation of the NF-κB/NFATc1 signaling pathway[J]. Am J Transl Res, 2017, 9(11): 5022-5030. |
| [37] | Li HW, Li Q, Guo T, et al. LncRNA CRNDE triggers inflammation through the TLR3-NF-κB-Cytokine signaling pathway[J]. Tumour Biol, 2017, 39(6): 1010428317703821. |
| [38] | Zhou CK, Gao J, Ji HY, et al. Benzoylaconine modulates LPS-induced responses through inhibition of toll-like receptor-mediated NF-κB and MAPK signaling in RAW264.7 cells[J]. Inflammation, 2021, 44(5): 2018-2032. |
| [39] | Zhou HY, Simion V, Pierce JB, et al. LncRNA-MAP3K4 regulates vascular inflammation through the p38 MAPK signaling pathway and cis-modulation of MAP3K4[J]. FASEB J, 2021, 35(1): e21133. |
| [40] |
Barrio L, Roman-Garcia S, Diaz-Mora E, et al. B cell development and T-dependent antibody response are regulated by p38γ and p38δ[J]. Front Cell Dev Biol, 2020, 8: 189.
doi: 10.3389/fcell.2020.00189 pmid: 32266269 |
| [41] | Sharma A, Tirpude NV, Kumari M, et al. Rutin prevents inflammation-associated colon damage via inhibiting the p38/MAPKAPK2 and PI3K/Akt/GSK3β/NF-κB signalling axes and enhancing splenic Tregs in DSS-induced murine chronic colitis[J]. Food Funct, 2021, 12(18): 8492-8506. |
| [42] | Ohto U. Activation and regulation mechanisms of NOD-like receptors based on structural biology[J]. Front Immunol, 2022, 13: 953530. |
| [43] |
Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling[J]. Nat Rev Immunol, 2016, 16(7): 407-420.
doi: 10.1038/nri.2016.58 pmid: 27291964 |
| [44] | Wang XZ, Liu MC, Geng N, et al. Staphylococcus aureus mediates pyroptosis in bovine mammary epithelial cell via activation of NLRP3 inflammasome[J]. Vet Res, 2022, 53(1): 10. |
| [45] | Ma MR, Pei YF, Wang XX, et al. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-κB/NLRP3 inflammasome pathway[J]. Cell Prolif, 2019, 52(1): e12525. |
| [46] | Wang DH, Höing S, Patterson HC, et al. Inflammation in mice ectopically expressing human Pyogenic Arthritis, Pyoderma Gangrenosum, and Acne(PAPA)Syndrome-associated PSTPIP1 A230T mutant proteins[J]. J Biol Chem, 2013, 288(7): 4594-4601. |
| [47] |
Huang W, Li YY, Zhang C, et al. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer[J]. J Cell Mol Med, 2020, 24(23): 13949-13960.
doi: 10.1111/jcmm.16003 pmid: 33094561 |
| [48] |
Kumar N, Sharma N, Mehan S. Connection between JAK/STAT and PPARγ signaling during the progression of multiple sclerosis: insights into the modulation of T-cells and immune responses in the brain[J]. Curr Mol Pharmacol, 2021, 14(5): 823-837.
doi: 10.2174/1874467214666210301121432 pmid: 33645493 |
| [49] | Zhuo Q, Wei L, Yin XT, et al. LncRNA ZNF667-AS1 alleviates rheumatoid arthritis by sponging miR-523-3p and inactivating the JAK/STAT signalling pathway[J]. Autoimmunity, 2021, 54(7): 406-414. |
| [50] |
Szydłowski M, Dębek S, Prochorec-Sobieszek M, et al. PIM kinases promote survival and immune escape in primary mediastinal large B-cell lymphoma through modulation of JAK-STAT and NF-κB activity[J]. Am J Pathol, 2021, 191(3): 567-574.
doi: 10.1016/j.ajpath.2020.12.001 pmid: 33307035 |
| [51] |
Xu ZH, Gwin KA, Li YL, et al. Developmental stage-specific effects of Pim-1 dysregulation on murine bone marrow B cell development[J]. BMC Immunol, 2016, 17(1): 16.
doi: 10.1186/s12865-016-0152-1 pmid: 27287229 |
| [52] | de Vries M, Heijink IH, Gras R, et al. Pim1 kinase protects airway epithelial cells from cigarette smoke-induced damage and airway inflammation[J]. Am J Physiol Lung Cell Mol Physiol, 2014, 307(3): L240-L251. |
| [1] | 金博阳, 秦仕宇, 张明达, 李倩倩, 文静, 沈秀丽, 杜志强. 小龙虾prx 6基因在对抗金黄色葡萄球菌感染中的分子作用机制研究[J]. 生物技术通报, 2024, 40(7): 314-322. |
| [2] | 张清兰, 张亚冉, 鞠志花, 王秀革, 肖遥, 王金鹏, 魏晓超, 高亚平, 白福恒, 王洪程. 牛TARDBP基因核心启动子鉴定与转录调控分析[J]. 生物技术通报, 2024, 40(4): 306-318. |
| [3] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [4] | 陈晓萌, 张雪静, 张欢, 张宝江, 苏艳. 重组牛乳源金黄色葡萄球菌GapC蛋白优势B细胞抗原表位的预测和筛选[J]. 生物技术通报, 2023, 39(5): 306-313. |
| [5] | 李彦霞, 王晋鹏, 冯芬, 包斌武, 董益闻, 王兴平, 罗仍卓么. 大肠杆菌型奶牛乳房炎对产奶性状相关基因表达的影响[J]. 生物技术通报, 2023, 39(2): 274-282. |
| [6] | 程深伟, 张克强, 梁军锋, 刘福元, 郜兴亮, 杜连柱. 畜禽养殖粪污中典型致病菌的三重微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2022, 38(9): 271-280. |
| [7] | 李宇航, 王兴平, 杨箭, 罗仍卓么, 任倩倩, 魏大为, 马云. miR-665在奶牛乳腺上皮细胞炎症中的表达及功能分析[J]. 生物技术通报, 2022, 38(5): 159-168. |
| [8] | 岑潇龙, 雷曦, 马诗云, 陈倩茹, 黄遵锡, 周峻沛, 张蕊. 金黄色葡萄球菌透明质酸裂解酶HylS的异源表达与特性研究[J]. 生物技术通报, 2022, 38(1): 157-167. |
| [9] | 陈立杰, 杨帆, 范海燕, 赵迪, 王媛媛, 朱晓峰, 刘晓宇, 段玉玺. 非编码RNA在生防菌-植物线虫-寄主互作中的研究进展[J]. 生物技术通报, 2021, 37(7): 65-70. |
| [10] | 王晋鹏, 罗仍卓么, 王兴平, 杨箭, 贾立, 马云, 魏大为. 奶牛乳腺炎治疗及抗炎分子机制的研究进展[J]. 生物技术通报, 2021, 37(12): 212-219. |
| [11] | 蒋成辉, 曾巧英, 王萌, 潘阳阳, 刘旭明, 尚天甜. CRISPR/Cas9构建srtA基因敲除的金黄色葡萄球菌[J]. 生物技术通报, 2020, 36(9): 253-265. |
| [12] | 张萌, 罗芳, 王敏, 武彦泽, 王俊奎, 和东迁, 陈丽尧, 陶金忠. 奶牛分娩后早期血浆代谢物变化研究[J]. 生物技术通报, 2020, 36(6): 191-199. |
| [13] | 吴家劲, 朱森林, 周密, 孙会增. 奶牛瘤胃微生物研究进展和趋势[J]. 生物技术通报, 2020, 36(2): 27-38. |
| [14] | 胡启超, 罗仍卓么, 魏大为, 杨箭, 贾立, 王兴平, 马云. 固有免疫相关编码基因在奶牛乳腺炎调节中的研究进展[J]. 生物技术通报, 2020, 36(12): 239-246. |
| [15] | 张萌, 刘国林, 李向龙, 陈永宏, 白玲荣, 罗芳, 李亚超, 陶金忠. 围产前期添加山楂和黄芪混合物对奶牛血浆代谢组的影响[J]. 生物技术通报, 2019, 35(8): 127-137. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||