








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 306-318.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1048
张清兰1,2( ), 张亚冉2,3(
), 张亚冉2,3( ), 鞠志花2,3, 王秀革2,3, 肖遥2,3, 王金鹏2,3, 魏晓超2,3, 高亚平2,3, 白福恒4, 王洪程1(
), 鞠志花2,3, 王秀革2,3, 肖遥2,3, 王金鹏2,3, 魏晓超2,3, 高亚平2,3, 白福恒4, 王洪程1( )
)
收稿日期:2023-11-09
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
张亚冉,女,博士,助理研究员,研究方向:奶牛分子育种;E-mail: zhang_ya_ran@126.com;作者简介:张清兰,女,硕士研究生,研究方向:奶牛分子育种;E-mail: 1400304672@qq.com
基金资助:
ZHANG Qing-lan1,2( ), ZHANG Ya-ran2,3(
), ZHANG Ya-ran2,3( ), JU Zhi-hua2,3, WANG Xiu-ge2,3, XIAO Yao2,3, WANG Jin-peng2,3, WEI Xiao-chao2,3, GAO Ya-ping2,3, BAI Fu-heng4, WANG Hong-cheng1(
), JU Zhi-hua2,3, WANG Xiu-ge2,3, XIAO Yao2,3, WANG Jin-peng2,3, WEI Xiao-chao2,3, GAO Ya-ping2,3, BAI Fu-heng4, WANG Hong-cheng1( )
)
Received:2023-11-09
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】为了解牛TARDBP基因结构、功能及启动子活性区域,分析该基因的转录调控机制。【方法】通过PCR技术克隆牛TARDBP基因编码区全长序列。应用生物信息学软件对牛TARDBP蛋白性质和结构进行分析。通过实时荧光定量PCR(RT-qPCR)技术检测TARDBP基因在泌乳期荷斯坦奶牛不同组织中的表达情况。应用5' RACE技术确定牛TARDBP基因的转录起始位点,PCR技术克隆牛TARDBP基因启动子区和5'端系列缺失片段,并运用生物信息学软件对牛TARDBP基因启动子区域CpG岛和潜在转录因子结合位点进行预测,双荧光素酶报告系统确定该基因的核心启动子区域。【结果】牛TARDBP基因在包括乳腺在内的多个组织中均有表达。TARDBP蛋白是一种水溶性非分泌蛋白,存在RNA识别结构域和DNA结合位点。TARDBP基因启动子区存在2个CpG岛和大量转录因子结合位点包括SP1、PPARA、PPARD、PPARG、SREBF1等。双荧光素酶活性分析结果显示牛TARDBP的核心启动子区位于-476 - -149之间,该区域包含Sp1、PPARG、PPARD、SREBF1结合元件,但不存在TATA-box。【结论】牛TARDBP基因在奶牛多个组织中均有表达。-476 - -149片段为牛TARDBP基因的核心启动子区,且该区域中存在与乳脂合成和分泌相关的转录因子结合位点。
张清兰, 张亚冉, 鞠志花, 王秀革, 肖遥, 王金鹏, 魏晓超, 高亚平, 白福恒, 王洪程. 牛TARDBP基因核心启动子鉴定与转录调控分析[J]. 生物技术通报, 2024, 40(4): 306-318.
ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene[J]. Biotechnology Bulletin, 2024, 40(4): 306-318.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product length/bp | 退火温度 Annealing temperature/℃ | 用途 Usage |
|---|---|---|---|---|
| TARDBP-F | ATGTCTGAATATATTCGGGTAACCG | 1 245 | 64.4 | CDS区扩增 |
| TARDBP-R | CTACATTCCCCAGCCAGAAGATTTA | |||
| TARDBP-GSP | CCGTGAAGCGAACGAAGCCAAACC | 496 | 68 | 5'RACE第一轮PCR |
| TARDBP -NGSP | AGATTCCCCCAGCCAGCATCGG | 230 | 5'RACE第二轮PCR | |
| pTARDBP-F | GCAAAGGGATAGAGGGAA | 1 954 | 55 | 启动子区扩增 |
| pTARDBP-R | CAGGGCGTCCTTAGTGAG | |||
| pGL3-P1 | CGG GGTACCGCAAAGGGATAGAGGGAA | 1 954 | 55 | 不同缺失片段扩增 |
| pGL3-P2 | CGG GGTACCGTTCAAACTACCGCACAATG | 1 498 | ||
| pGL3-P3 | CGG GGTACCTAGGGTGGCAAAGAGTTGG | 1 179 | ||
| pGL3-P4 | CGG GGTACCGAGAAGGAAATGGCAACCC | 854 | ||
| pGL3-P5 | CGG GGTACCAATGGCTTGGGCTCAACAG | 615 | ||
| pGL3-P6 | CGG GGTACCTATAGCCTTCAAGTTCACTCCC | 288 | ||
| pGL3-R1 | CCC AAGCTTCAGGGCGTCCTTAGTGAG | |||
| TARDBP-q-F | GTTTGGCTTCGTTCGCTTCA | 164 | 60 | RT-qPCR |
| TARDBP-q-R | CCTCTGTACAACGCCCAACA | |||
| β-actin-q-F | CATCGGCAATGAGCGGTTCC | 147 | ||
| β-actin-q-R | ACCGTGTTGGCGTAGAGGTC |
表1 牛TARDBP基因引物序列信息
Table 1 Sequence information of bovine TARDBP gene primer
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product length/bp | 退火温度 Annealing temperature/℃ | 用途 Usage |
|---|---|---|---|---|
| TARDBP-F | ATGTCTGAATATATTCGGGTAACCG | 1 245 | 64.4 | CDS区扩增 |
| TARDBP-R | CTACATTCCCCAGCCAGAAGATTTA | |||
| TARDBP-GSP | CCGTGAAGCGAACGAAGCCAAACC | 496 | 68 | 5'RACE第一轮PCR |
| TARDBP -NGSP | AGATTCCCCCAGCCAGCATCGG | 230 | 5'RACE第二轮PCR | |
| pTARDBP-F | GCAAAGGGATAGAGGGAA | 1 954 | 55 | 启动子区扩增 |
| pTARDBP-R | CAGGGCGTCCTTAGTGAG | |||
| pGL3-P1 | CGG GGTACCGCAAAGGGATAGAGGGAA | 1 954 | 55 | 不同缺失片段扩增 |
| pGL3-P2 | CGG GGTACCGTTCAAACTACCGCACAATG | 1 498 | ||
| pGL3-P3 | CGG GGTACCTAGGGTGGCAAAGAGTTGG | 1 179 | ||
| pGL3-P4 | CGG GGTACCGAGAAGGAAATGGCAACCC | 854 | ||
| pGL3-P5 | CGG GGTACCAATGGCTTGGGCTCAACAG | 615 | ||
| pGL3-P6 | CGG GGTACCTATAGCCTTCAAGTTCACTCCC | 288 | ||
| pGL3-R1 | CCC AAGCTTCAGGGCGTCCTTAGTGAG | |||
| TARDBP-q-F | GTTTGGCTTCGTTCGCTTCA | 164 | 60 | RT-qPCR |
| TARDBP-q-R | CCTCTGTACAACGCCCAACA | |||
| β-actin-q-F | CATCGGCAATGAGCGGTTCC | 147 | ||
| β-actin-q-R | ACCGTGTTGGCGTAGAGGTC |
| 软件名Software name | 网址 Website | 用途 Purpose |
|---|---|---|
| NCBI | https://www.ncbi.nlm.nih.gov/ | 氨基酸序列 |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 保守结构域分析 | |
| DNAMAN9.0软件 | 同源性比对 | |
| MEGA11.0软件 | 构建基因系统进化树 | |
| ExPASy | https://web.expasy.org/protscale/ | 理化性质分析 |
| TMHMM | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 蛋白跨膜结构域预测 |
| SignalP5.0 | https://services.healthtech.dtu.dk/service.php?Signal P-5.0 | 蛋白信号肽预测 |
| NetPhos-3.1 | http://www.cbs.dtu.dk/services/NetPhos/ | 蛋白磷酸化位点分析 |
| NPS@SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl | 蛋白质二级结构预测 |
| SWISS-MODEL | https://swissmodel.expasy.org/ | 蛋白质三级结构预测 |
| Meth Primer | http://www.urogene.org/methprimer/ | CpG岛预测 |
| AnimalTFDB4 | bioinfo.life.hust.edu.cn/AnimalTFDB4 | 转录因子结合位点预测 |
| TRANSFAC2.0 | https://genexplain.com/transfac-2-0/ |
表2 生物信息学分析用软件
Table 2 Software for bioinformatics analysis
| 软件名Software name | 网址 Website | 用途 Purpose |
|---|---|---|
| NCBI | https://www.ncbi.nlm.nih.gov/ | 氨基酸序列 |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 保守结构域分析 | |
| DNAMAN9.0软件 | 同源性比对 | |
| MEGA11.0软件 | 构建基因系统进化树 | |
| ExPASy | https://web.expasy.org/protscale/ | 理化性质分析 |
| TMHMM | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 蛋白跨膜结构域预测 |
| SignalP5.0 | https://services.healthtech.dtu.dk/service.php?Signal P-5.0 | 蛋白信号肽预测 |
| NetPhos-3.1 | http://www.cbs.dtu.dk/services/NetPhos/ | 蛋白磷酸化位点分析 |
| NPS@SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl | 蛋白质二级结构预测 |
| SWISS-MODEL | https://swissmodel.expasy.org/ | 蛋白质三级结构预测 |
| Meth Primer | http://www.urogene.org/methprimer/ | CpG岛预测 |
| AnimalTFDB4 | bioinfo.life.hust.edu.cn/AnimalTFDB4 | 转录因子结合位点预测 |
| TRANSFAC2.0 | https://genexplain.com/transfac-2-0/ |
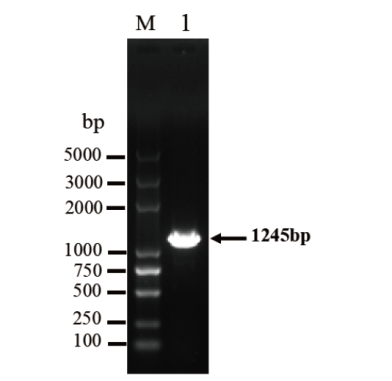
图1 牛TARDBP基因CDS区PCR产物 M:DL5000 DNA marker;1:牛TARDBP基因CDS区
Fig. 1 PCR product of the bovine TARDBP gene CDS region M: DL5000 DNA marker. 1: CDS region of bovine TARDBP gene
| 物种 Species | 登录号 GenBank No. | 氨基酸序列相似性Similarity/% |
|---|---|---|
| 牛 Bos taurus | NP_001039950.2 | |
| 山羊 Capra hircus | XP_005690745.1 | 100 |
| 绵羊 Ovis aries | XP_027831445.1 | 100 |
| 犬 Canis lupus familiaris | XP_038516303.1 | 99.76 |
| 猪 Sus scrofa | XP_020950991.1 | 99.52 |
| 家猫 Felis catus | XP_003989534.1 | 99.03 |
| 人 Homo sapiens | NP_031401.1 | 97.58 |
| 苏门答腊猩猩Pongo abelii | NP_001127597.1 | 97.34 |
| 原鸡 Gallus gallus | NP_001026049.2 | 96.86 |
| 热带爪蟾 Xenopus tropicalis | NP_001231687.1 | 74.10 |
| 小鼠 Mus musculus | NP_001003898.1 | 67.39 |
| 大鼠 Rattus norvegicus | NP_001011979.1 | 66.67 |
表3 牛与不同物种TARDBP氨基酸序列比对
Table 3 Amino acid sequence comparison of TARDBP between cattle and different species
| 物种 Species | 登录号 GenBank No. | 氨基酸序列相似性Similarity/% |
|---|---|---|
| 牛 Bos taurus | NP_001039950.2 | |
| 山羊 Capra hircus | XP_005690745.1 | 100 |
| 绵羊 Ovis aries | XP_027831445.1 | 100 |
| 犬 Canis lupus familiaris | XP_038516303.1 | 99.76 |
| 猪 Sus scrofa | XP_020950991.1 | 99.52 |
| 家猫 Felis catus | XP_003989534.1 | 99.03 |
| 人 Homo sapiens | NP_031401.1 | 97.58 |
| 苏门答腊猩猩Pongo abelii | NP_001127597.1 | 97.34 |
| 原鸡 Gallus gallus | NP_001026049.2 | 96.86 |
| 热带爪蟾 Xenopus tropicalis | NP_001231687.1 | 74.10 |
| 小鼠 Mus musculus | NP_001003898.1 | 67.39 |
| 大鼠 Rattus norvegicus | NP_001011979.1 | 66.67 |
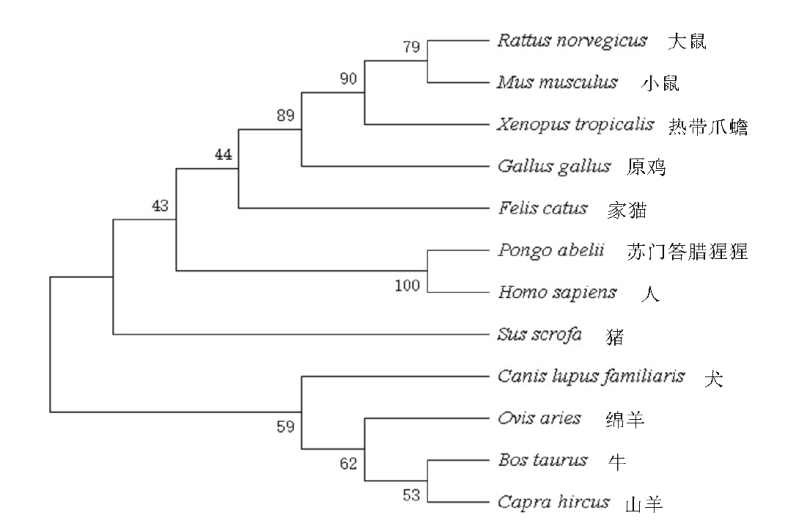
图2 TARDBP基因系统进化树 树枝上方数字是>50 %的自展值(1 000次重复)
Fig. 2 Phylogenetic tree of TARDBP gene The number on the branch is >50% of the self-explanatory value(1 000 repetitions)
| 氨基酸 Amino acid | 数目 Number | 比例Perc-entage/% | 氨基酸 Amino acid | 数目 Number | 比例Perc- entage/% |
|---|---|---|---|---|---|
| Ala(A) | 27 | 6.5% | Leu(L) | 20 | 4.8% |
| Arg(R) | 20 | 4.8% | Lys(K) | 20 | 4.8% |
| Asn(N) | 27 | 6.5% | Met(M) | 17 | 4.1% |
| Asp(D) | 22 | 5.3% | Phe(F) | 22 | 5.3% |
| Cys(C) | 7 | 1.7% | Pro(P) | 17 | 4.1% |
| Gln(Q) | 24 | 5.8% | Ser(S) | 41 | 9.9% |
| Glu(E) | 20 | 4.8% | Thr(T) | 15 | 3.6% |
| Gly(G) | 55 | 13.3% | Trp(W) | 6 | 1.4% |
| His(H) | 5 | 1.2% | Tyr(Y) | 6 | 1.9% |
| Ile(I) | 14 | 3.4% | Val(V) | 8 | 6.5% |
表4 牛TARDBP蛋白氨基酸组成
Table 4 Amino acid composition of bovine TARDBP protein
| 氨基酸 Amino acid | 数目 Number | 比例Perc-entage/% | 氨基酸 Amino acid | 数目 Number | 比例Perc- entage/% |
|---|---|---|---|---|---|
| Ala(A) | 27 | 6.5% | Leu(L) | 20 | 4.8% |
| Arg(R) | 20 | 4.8% | Lys(K) | 20 | 4.8% |
| Asn(N) | 27 | 6.5% | Met(M) | 17 | 4.1% |
| Asp(D) | 22 | 5.3% | Phe(F) | 22 | 5.3% |
| Cys(C) | 7 | 1.7% | Pro(P) | 17 | 4.1% |
| Gln(Q) | 24 | 5.8% | Ser(S) | 41 | 9.9% |
| Glu(E) | 20 | 4.8% | Thr(T) | 15 | 3.6% |
| Gly(G) | 55 | 13.3% | Trp(W) | 6 | 1.4% |
| His(H) | 5 | 1.2% | Tyr(Y) | 6 | 1.9% |
| Ile(I) | 14 | 3.4% | Val(V) | 8 | 6.5% |
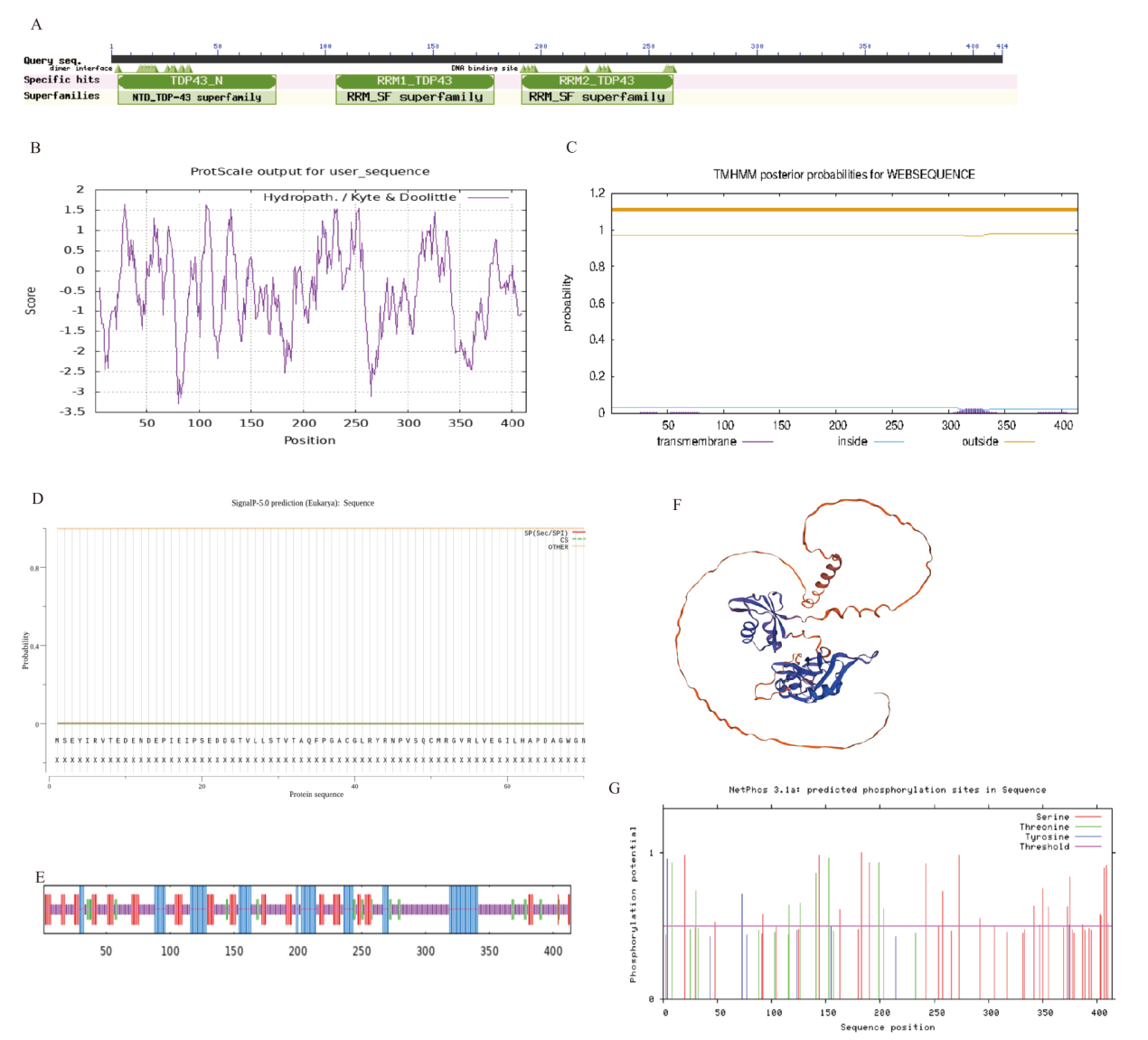
图3 TARDBP蛋白结构分析 A:牛TARDBP蛋白保守结构域预测;B:牛TARDBP蛋白疏水性预测;C:牛TARDBP蛋白跨膜结构预测;D:牛TARDBP蛋白信号肽预测;E:牛TARDBP蛋白二级结构预测;F:牛TARDBP蛋白三级结构预测;G:牛TARDBP蛋白磷酸化位点预测
Fig. 3 Analysis of TARDBP protein structure A: Prediction of bovine TARDBP protein conserved domain. B: Prediction of bovine TARDBP protein hydrophobicity. C: Prediction of bovine TARDBP protein transmembrane structure. D: Prediction of bovine TARDBP protein signal peptide. E: Prediction of secondary structure of bovine TARDBP protein. F: Tertiary structure prediction of bovine TARDBP protein. G: Prediction of phosphorylation site of bovine TARDBP protein
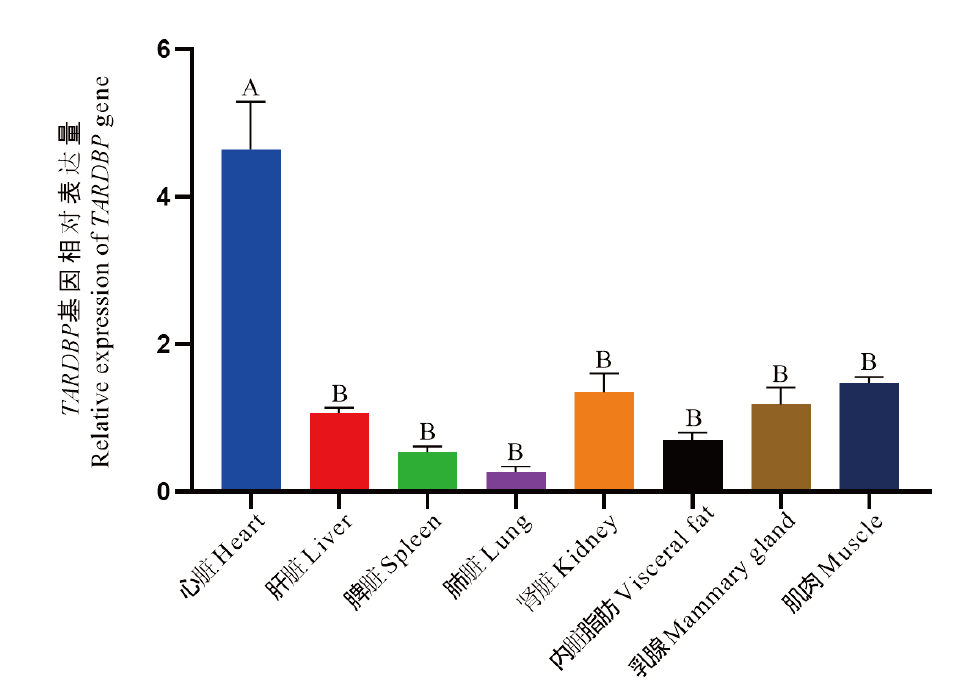
图4 牛TARDBP基因在不同组织的表达分析 相同大写字母表示差异不显著(P>0.05),不同大写字母表示差异极显著(P<0.01)
Fig. 4 Analysis of bovine TARDBP gene expression in different tissues The same capital letter indicates that the difference is not significant(P>0.05), different capital letters indicate significant differences(P<0.01)
图5 牛TARDBP基因5' RACE扩增电泳图 M:DL5000 DNA marker;1:第一轮PCR产物;2:第二轮PCR产物
Fig. 5 5' RACE amplification electrophoresis of bovine TARDBP gene M: DL5000 DNA marker. 1: First round PCR products. 2: Second round PCR product
图7 牛TARDBP基因启动子区PCR产物 M:DL5000 DNA marker;1:牛TARDBP基因 5'侧翼区-1815/+139 片段
Fig. 7 PCR products from the promoter region of bovine TARDBP gene M: DL5000 DNA marker. 1: The 5' flanking region(-1815/+139)of bovine TARDBP gene
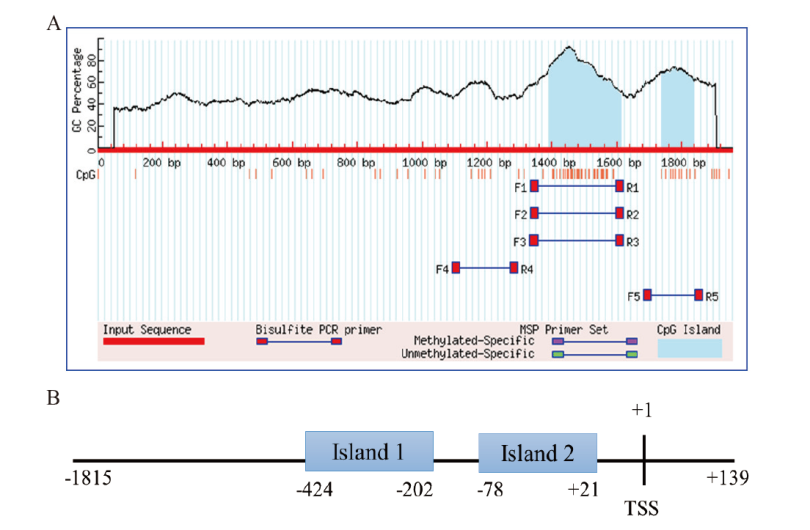
图8 牛TARDBP基因5'调控区CpG island 预测结果 A:MethPrimer 软件预测结果;B:CpG islands 在序列中的定位
Fig. 8 Prediction results of CpG island in the 5' regulatory region of bovine TARDBP gene A: The predict result by MethPrimer software. B: The specific location of CpG islands

图9 牛TARDBP基因启动子潜在的转录因子结合位点预测结果 +1表示转录起始位点;方框表示转录因子结合位点;重叠的转录因子结合位点区域用不同颜色的框表示
Fig. 9 Prediction of potential transcription factor binding sites of bovine TARDBP gene promoter +1 indicates transcription initiation site; letters in in box indicates transcription factor binding sites. Overlapping regions of transcription factor binding sites are represented by different colored boxes

图10 牛TARDBP基因启动子不同缺失片段的扩增与载体构建 A:牛TARDBP基因启动子逐段缺失片段扩增电泳图,M:DL5000 DNA marker,1-6:加双酶切位点牛TARDBP基因启动子逐段缺失片段;B:牛TARDBP基因启动子双荧光素报告重组质粒双酶切鉴定电泳图,M:DL5000 DNA marker,1-6:重组质粒
Fig. 10 Amplification and vector construction of different deletion fragments of bovine TARDBP gene promoter A: Electrophoretic amplification of bovine TARDBP gene promoter fragment by fragment deletion, M: DL5000 DNA marker, 1-6: add the missing fragments of bovine TARDBP gene promoter at double restriction sites. B: Electrophoresis of bovine TARDBP gene promoter double fluorescein reporter recombinant plasmid double enzyme digestion identification, M: DL5000 DNA marker, 1-6: recombinant plasmid
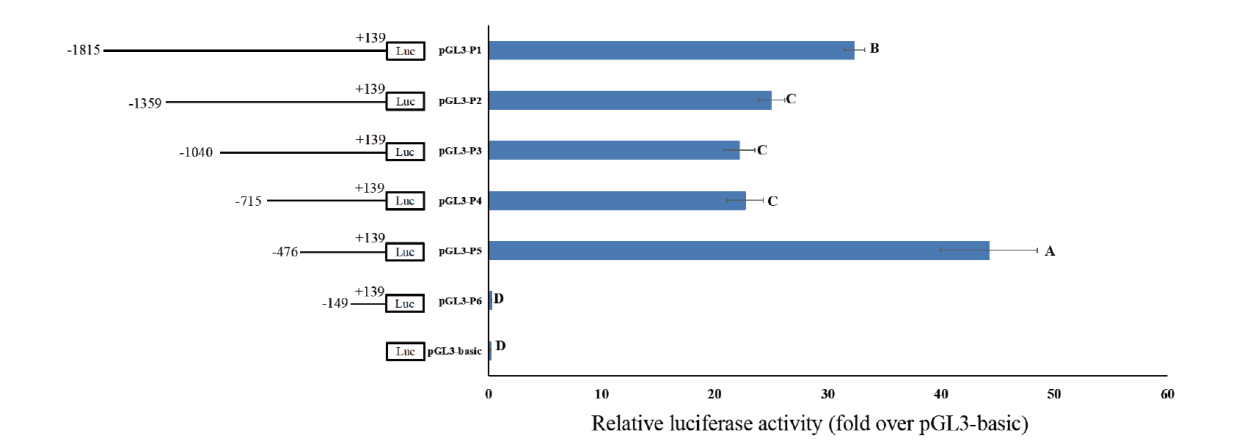
图11 牛TARDBP基因启动子在牛乳腺上皮细胞中的活性分析 不同字母表示差异显著(P<0.05)
Fig. 11 Activity analysis of bovine TARDBP gene promoter in bovine mammary epithelial cells Different letters indicate significant differences(P<0.05)
| [1] |
Chalupa-Krebzdak S, Long CJ, Bohrer BM. Nutrient density and nutritional value of milk and plant-based milk alternatives[J]. Int Dairy J, 2018, 87: 84-92.
doi: 10.1016/j.idairyj.2018.07.018 URL |
| [2] | Wijesinha-Bettoni R, Burlingame B. Milk and dairy product composition[J]. Milk and dairy products in human nutrition, 2013: 41-102. |
| [3] | Fox PF, Mcsweeney PLH, Paul LH. Dairy chemistry and biochemistry[M]. London: Chapman & Hall,1999. |
| [4] | Samková E, Špička J, Pešek M, et al. Animal factors affecting fatty acid composition of cow milk fat: a review[J]. S Afr N J Anim Sci, 2012, 42(2): 83-100. |
| [5] |
蒋秋斐, 蔡正云, 黄增文, 等. 奶牛乳脂性状候选基因EEF1D突变位点功能分析[J]. 浙江农业学报, 2020, 32(7): 1155-1159.
doi: 10.3969/j.issn.1004-1524.2020.07.03 |
| Jiang QF, Cai ZY, Huang ZW, et al. Functional analysis of EEF1D mutation site in dairy cow milk fat traits candidate gene[J]. Acta Agric Zhejiangensis, 2020, 32(7): 1155-1159. | |
| [6] | Xu B, Gerin I, Miao HZ, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity[J]. PLoS One, 2010, 5(12): e14199. |
| [7] | Chhangani D, Martín-Peña A, Rincon-Limas DE. Molecular, functional, and pathological aspects of TDP-43 fragmentation[J]. iScience, 2021, 24(5): 102459. |
| [8] |
Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function[J]. Curr Opin Cell Biol, 1999, 11(3): 363-371.
pmid: 10395553 |
| [9] |
Lukavsky PJ, Daujotyte D, Tollervey JR, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43[J]. Nat Struct Mol Biol, 2013, 20(12): 1443-1449.
doi: 10.1038/nsmb.2698 pmid: 24240615 |
| [10] |
Weskamp K, Barmada SJ. TDP43 and RNA instability in amyotrophic lateral sclerosis[J]. Brain Res, 2018, 1693(Pt A): 67-74.
doi: S0006-8993(18)30023-4 pmid: 29395044 |
| [11] |
Kuo PH, Chiang CH, Wang YT, et al. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids[J]. Nucleic Acids Res, 2014, 42(7): 4712-4722.
doi: 10.1093/nar/gkt1407 URL |
| [12] |
Buratti E, Dörk T, Zuccato E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping[J]. EMBO J, 2001, 20(7): 1774-1784.
doi: 10.1093/emboj/20.7.1774 pmid: 11285240 |
| [13] |
Baralle M, Buratti E, Baralle FE. The role of TDP-43 in the pathogenesis of ALS and FTLD[J]. Biochem Soc Trans, 2013, 41(6): 1536-1540.
doi: 10.1042/BST20130186 URL |
| [14] |
Zhao LM, Li LL, Xu HB, et al. TDP-43 is required for mammary gland repopulation and proliferation of mammary epithelial cells[J]. Stem Cells Dev, 2019, 28(14): 944-953.
doi: 10.1089/scd.2019.0011 pmid: 31062657 |
| [15] | Zhao LM, Ke H, Xu HB, et al. TDP-43 facilitates milk lipid secretion by post-transcriptional regulation of Btn1a1 and Xdh[J]. Nat Commun, 2020, 11(1): 341. |
| [16] | Gowardhan A, Spoon H, Riechers DA, et al. The dual role of starbursts and active galactic nuclei in driving extreme molecular outflows[J]. ApJ, 2018, 859(1): 35. |
| [17] |
Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease[J]. Front Biosci, 2008, 13: 867-878.
doi: 10.2741/2727 URL |
| [18] |
Warraich ST, Yang S, Nicholson GA, et al. TDP-43: a DNA and RNA binding protein with roles in neurodegenerative diseases[J]. Int J Biochem Cell Biol, 2010, 42(10): 1606-1609.
doi: 10.1016/j.biocel.2010.06.016 pmid: 20601083 |
| [19] |
Stover CM, Lynch NJ, Hanson SJ, et al. Organization of the MASP2 locus and its expression profile in mouse and rat[J]. Mamm Genome, 2004, 15(11): 887-900.
pmid: 15672593 |
| [20] |
Ou SH, Wu F, Harrich D, et al. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs[J]. J Virol, 1995, 69(6): 3584-3596.
doi: 10.1128/JVI.69.6.3584-3596.1995 pmid: 7745706 |
| [21] | 郑敏. 家猪全基因组甲基化时空变化以及重复元件PRE1甲基化功能研究[D]. 南昌: 江西农业大学, 2022. |
| Zheng M. Study on the spatiotemporal variations of pig methylomes and the function of methylated PRE1[D]. Nanchang: Jiangxi Agricultural University, 2022. | |
| [22] | Oh YM, Mahar M, Ewan EE, et al. Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration[J]. Proc Natl Acad Sci USA, 2018, 115(52): E12417-E12426. |
| [23] | Xiang Y, Chen QX, Li QB, et al. The expression level of chicken telomerase reverse transcriptase in tumors induced by ALV-J is positively correlated with methylation and mutation of its promoter region[J]. Vet Res, 2022, 53(1): 49. |
| [24] |
Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation[J]. Nat Rev Genet, 2012, 13(4): 233-245.
doi: 10.1038/nrg3163 pmid: 22392219 |
| [25] |
Kadonaga JT, Carner KR, Masiarz FR, et al. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain[J]. Cell, 1987, 51(6): 1079-1090.
doi: 10.1016/0092-8674(87)90594-0 pmid: 3319186 |
| [26] |
Song J, Ugai H, Nakata-Tsutsui H, et al. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements[J]. Int J Mol Med, 2003, 11(5): 547-553.
pmid: 12684688 |
| [27] | 杨洋, 周子薇, 张京一, 等. 奶牛SP1基因结构及对乳脂合成功能的初步分析[J]. 畜牧兽医学报, 2022, 53(9): 2970-2981. |
| Yang Y, Zhou ZW, Zhang JY, et al. Analysison SP1 gene structure and Its functionon milk fat synthesisin holstein dairy cows[J]. Acta Veterinaria Et Zootechnica Sinica, 2022, 53(9):2970-2981. | |
| [28] |
Zhu JJ, Luo J, Xu HF, et al. Short communication: altered expression of specificity protein 1 impairs milk fat synthesis in goat mammary epithelial cells[J]. J Dairy Sci, 2016, 99(6): 4893-4898.
doi: S0022-0302(16)30086-8 pmid: 26995134 |
| [29] |
Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle[J]. BMC Genomics, 2008, 9: 366.
doi: 10.1186/1471-2164-9-366 pmid: 18671863 |
| [30] |
Yu K, Shi HB, Luo J, et al. PPARG modulated lipid accumulation in dairy GMEC via regulation of ADRP gene[J]. J Cell Biochem, 2015, 116(1): 192-201.
doi: 10.1002/jcb.v116.1 URL |
| [31] |
Kadegowda AKG, Bionaz M, Piperova LS, et al. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents[J]. J Dairy Sci, 2009, 92(9): 4276-4289.
doi: 10.3168/jds.2008-1932 pmid: 19700688 |
| [32] |
Shi HB, Luo J, Yao DW, et al. Peroxisome proliferator-activated receptor-γ stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase[J]. J Dairy Sci, 2013, 96(12): 7844-7853.
doi: 10.3168/jds.2013-7105 URL pmid: 24119817 |
| [33] |
Shi HB, Zhao WS, Luo J, et al. Peroxisome proliferator-activated receptor γ1 and γ2 isoforms alter lipogenic gene networks in goat mammary epithelial cells to different extents[J]. J Dairy Sci, 2014, 97(9): 5437-5447.
doi: 10.3168/jds.2013-7863 pmid: 25022676 |
| [34] |
Shi HB, Zhang CH, Xu ZA, et al. Peroxisome proliferator-activated receptor delta regulates lipid droplet formation and transport in goat mammary epithelial cells[J]. J Dairy Sci, 2018, 101(3): 2641-2649.
doi: S0022-0302(18)30014-6 pmid: 29331469 |
| [35] |
Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor[J]. Cell, 1997, 89(3): 331-340.
doi: 10.1016/s0092-8674(00)80213-5 pmid: 9150132 |
| [36] | Zhao X, Li J, Zhao SY, et al. Regulation of bta-miRNA29d-3p on lipid accumulation via GPAM in bovine mammary epithelial cells[J]. Agriculture, 2023, 13(2): 501. |
| [37] |
Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism[J]. Trends Endocrinol Metab, 2012, 23(2): 65-72.
doi: 10.1016/j.tem.2011.10.004 URL |
| [38] |
Cheng X, Li JY, Guo DL. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy[J]. Curr Top Med Chem, 2018, 18(6): 484-493.
doi: 10.2174/1568026618666180523104541 pmid: 29788888 |
| [39] |
Xue LY, Qi HY, Zhang H, et al. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy[J]. Front Oncol, 2020, 10: 1510.
doi: 10.3389/fonc.2020.01510 pmid: 32974183 |
| [40] |
Xu HF, Luo J, Zhao WS, et al. Overexpression of SREBP1(sterol regulatory element binding protein 1)promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells[J]. J Dairy Sci, 2016, 99(1): 783-795.
doi: 10.3168/jds.2015-9736 pmid: 26601584 |
| [1] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [2] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [3] | 陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145. |
| [4] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [5] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [6] | 朱毅, 柳唐镜, 宫国义, 张洁, 王晋芳, 张海英. 西瓜ClPP2C3克隆及表达分析[J]. 生物技术通报, 2024, 40(1): 243-249. |
| [7] | 刘玉玲, 王梦瑶, 孙琦, 马利花, 朱新霞. 启动子RD29A对转雪莲SikCDPK1基因烟草抗逆性的影响[J]. 生物技术通报, 2023, 39(9): 168-175. |
| [8] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [9] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [10] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [11] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [12] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| [13] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [14] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [15] | 邢媛, 宋健, 李俊怡, 郑婷婷, 刘思辰, 乔治军. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||