








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 260-269.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0279
王美玲1( ), 耿丽丽2, 房瑜1,2, 束长龙2, 张杰1,2(
), 耿丽丽2, 房瑜1,2, 束长龙2, 张杰1,2( )
)
收稿日期:2024-03-21
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
张杰,男,博士,研究员,研究方向:Bt菌株、基因资源挖掘与应用;E-mail: zhangjie05@caas.cn作者简介:王美玲,女,博士,讲师,研究方向:Bt菌株、基因资源挖掘与应用;E-mail: meilingw_123@163.com
基金资助:
WANG Mei-ling1( ), GENG Li-li2, FANG Yu1,2, SHU Chang-long2, ZHANG Jie1,2(
), GENG Li-li2, FANG Yu1,2, SHU Chang-long2, ZHANG Jie1,2( )
)
Received:2024-03-21
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】前期研究显示苏云金芽胞杆菌(Bacillus thuringiensis,Bt)4BM1菌株可诱导油菜产生对核盘菌(Sclerotinia sclerotiorum)的系统抗病性。探究其诱导系统抗性机制并分析其生防潜力,为油菜菌核病的生物防治提供菌株资源。【方法】采用实时荧光定量PCR和转录组测序技术分析根部施用4BM1菌株后,油菜叶片中防御相关基因的转录水平;结合4BM1菌株全基因组序列和antiSMASH 2.0软件预测其抗病相关次级代谢产物生物合成的基因簇;采用发酵液灌根的方式将4BM1菌株接种至油菜盆栽幼苗,分析其定殖能力和促生效果。【结果】4BM1菌株可激发油菜叶片产生过敏反应;油菜根部施用4BM1菌株,叶片中参与水杨酸、茉莉酸、乙烯信号和油菜素内酯生物合成途径等基因转录水平显著上调;4BM1菌株的染色体上预测到胞外多糖和其他13个抗病相关次级代谢产物合成的基因簇;4BM1菌株在油菜根际定殖的同时可促进油菜苗期植株的生长。【结论】4BM1菌株可作为防控油菜菌核病、促进油菜幼苗生长有潜力的生物防治资源。
王美玲, 耿丽丽, 房瑜, 束长龙, 张杰. 苏云金芽胞杆菌4BM1菌株对油菜菌核病的防治潜力[J]. 生物技术通报, 2024, 40(9): 260-269.
WANG Mei-ling, GENG Li-li, FANG Yu, SHU Chang-long, ZHANG Jie. Control Potential of Bacillus thuringiensis 4BM1 Strain to Sclerotiniose in Brassica campestris L.[J]. Biotechnology Bulletin, 2024, 40(9): 260-269.
| 功能 Function | 基因编号 Gene ID | 差异倍数的log值 log2Fold-change | 调控方式 Regulated | 描述 Description |
|---|---|---|---|---|
| Steroid-related genes | Bra029392* | 8.51283041 | Up | Squalene epoxidase |
| Bra031393 | 1.572668503 | Up | Sugar phosphate/phosphate translocator | |
| Ethylene-related genes | Bra004522 | 1.13850342 | Up | 26S protease regulatory subunit 8 |
| Bra002499* | 2.858810471 | Up | Concanavalin A-like lectin kinase-like protein | |
| Bra017656 | 2.011958293 | Up | Ethylene-responsive transcription factor | |
| Salicylic acid-related genes | Bra024318* | 1.115278771 | Up | U-box domain-containing protein 27 |
| Bra037520 | 1.415677575 | Up | Inorganic ion transport and metabolism | |
| Bra013123 | 2.021981527 | Up | Pathogenesis-related protein | |
| Bra013732 | 1.576452095 | Up | WRKY transcription factor | |
| Bra033158 | 1.751241545 | Up | WRKY transcription factor | |
| Bra008435 | 1.190167975 | Up | WRKY transcription factor | |
| Secondary metabolite biosynthesis-related genes | Bra040582 | 5.867390439 | Up | Sesquiterpenoid biosynthetic |
| Bra021965 | 1.653287924 | Up | Abscisic acid-hydroxylase 3 | |
| Chloroplast-related genes | Bra003004 | -6.601409988 | Down | Protochlorophyllide reductase A |
表1 4BM1菌株发酵液处理后油菜叶片中部分代谢作用相关的上调/下调基因列表
Table 1 List of up/down-regulated genes related to particular metabolisms in 4BM1-treated B. campestris leaves
| 功能 Function | 基因编号 Gene ID | 差异倍数的log值 log2Fold-change | 调控方式 Regulated | 描述 Description |
|---|---|---|---|---|
| Steroid-related genes | Bra029392* | 8.51283041 | Up | Squalene epoxidase |
| Bra031393 | 1.572668503 | Up | Sugar phosphate/phosphate translocator | |
| Ethylene-related genes | Bra004522 | 1.13850342 | Up | 26S protease regulatory subunit 8 |
| Bra002499* | 2.858810471 | Up | Concanavalin A-like lectin kinase-like protein | |
| Bra017656 | 2.011958293 | Up | Ethylene-responsive transcription factor | |
| Salicylic acid-related genes | Bra024318* | 1.115278771 | Up | U-box domain-containing protein 27 |
| Bra037520 | 1.415677575 | Up | Inorganic ion transport and metabolism | |
| Bra013123 | 2.021981527 | Up | Pathogenesis-related protein | |
| Bra013732 | 1.576452095 | Up | WRKY transcription factor | |
| Bra033158 | 1.751241545 | Up | WRKY transcription factor | |
| Bra008435 | 1.190167975 | Up | WRKY transcription factor | |
| Secondary metabolite biosynthesis-related genes | Bra040582 | 5.867390439 | Up | Sesquiterpenoid biosynthetic |
| Bra021965 | 1.653287924 | Up | Abscisic acid-hydroxylase 3 | |
| Chloroplast-related genes | Bra003004 | -6.601409988 | Down | Protochlorophyllide reductase A |
| 4BM1 | CDS | Bases | Gene | misc_RNA | rRNA | tRNA | Intergenetic region/% | GC/% | Gene GC/% | Intergenetic GC/% |
|---|---|---|---|---|---|---|---|---|---|---|
| 染色体Chromosome | 5529 | 5498518 | 5811 | 131 | 42 | 108 | 15.22 | 35.53 | 36.38 | 30.77 |
| 质粒1 Plasmid1 | 313 | 333074 | 318 | 5 | 0 | 0 | 21.78 | 33.37 | 34.29 | 30.06 |
| 质粒2 Plasmid2 | 205 | 218923 | 207 | 2 | 0 | 0 | 26.69 | 33.2 | 34.28 | 30.24 |
表2 4BM1菌株全基因组序列分析
Table 2 Analysis of genome-wide sequenc of 4BM1 strain
| 4BM1 | CDS | Bases | Gene | misc_RNA | rRNA | tRNA | Intergenetic region/% | GC/% | Gene GC/% | Intergenetic GC/% |
|---|---|---|---|---|---|---|---|---|---|---|
| 染色体Chromosome | 5529 | 5498518 | 5811 | 131 | 42 | 108 | 15.22 | 35.53 | 36.38 | 30.77 |
| 质粒1 Plasmid1 | 313 | 333074 | 318 | 5 | 0 | 0 | 21.78 | 33.37 | 34.29 | 30.06 |
| 质粒2 Plasmid2 | 205 | 218923 | 207 | 2 | 0 | 0 | 26.69 | 33.2 | 34.28 | 30.24 |
| 基因编号 Gene ID | 功能预测 Predicted function | 4F5菌株中预测的胞外多糖基因簇 Predicted eps gene cluster in Bt 4F5 strain | 4BM1与4F5之间的氨基酸序列相似性Amino acid identity between 4BM1 and 4F5/% |
|---|---|---|---|
| 4BM1_Chr1_00581 | EpsC | 4F5_Chr1_00600 | N/A |
| 4BM1_Chr1_00582 | UTP-glucose-1-phosphate uridylyltransferase(EC:2.7.7.9) | 4F5_Chr1_00601 | 94.5 |
| 4BM1_Chr1_00583 | Lipopolysaccharide synthesis sugar transferase | 4F5_Chr1_00602 | 94.9 |
| 4BM1_Chr1_00584 | Nucleotide sugar dehydrogenase | 4F5_Chr1_00603 | 23 |
| 4BM1_Chr1_00585 | Asparagine synthase | 4F5_Chr1_00604 | N/A |
| 4BM1_Chr1_00586 | Polysaccharide biosynthesis protein | 4F5_Chr1_00605 | N/A |
| 4BM1_Chr1_00587 | Glycosyltransferase | 4F5_Chr1_00606 | N/A |
| 4BM1_Chr1_00588 | Hypothetical protein | 4F5_Chr1_00607 | N/A |
| 4BM1_Chr1_00589 | Hypothetical protein | 4F5_Chr1_00608 | N/A |
| 4BM1_Chr1_00590 | Glycosyltransferases group 1 | 4F5_Chr1_00609 | N/A |
| 4BM1_Chr1_00591 | Glycosyltransferase, group 1 family protein | 4F5_Chr1_00610 | N/A |
| 4BM1_Chr1_00592 | Glycosyltransferase | 4F5_Chr1_00611 | N/A |
| 4BM1_Chr1_00593 | Glucose-1-phosphate thymidylyltransferase(EC:2.7.7.24) | 4F5_Chr1_00612 | N/A |
| 4BM1_Chr1_00594 | dTDP-4-dehydrorhamnose 3,5-epimerase(EC:5.1.3.13) | None | |
| 4BM1_Chr1_00595 | dTDP-glucose 4,6-dehydratase | None | |
| 4BM1_Chr1_00596 | dTDP-4-dehydrorhamnose reductase(EC:1.1.1.133) | None | |
| 4BM1_Chr1_00597 | Membrane-bound transcriptional regulator LytR | 4F5_Chr1_00613 | 88.5 |
| 4BM1_Chr1_00598 | EPSX protein | 4F5_Chr1_00614 | 89.9 |
| 4BM1_Chr1_00599 | UDP-glucose 4-epimerase GalE | 4F5_Chr1_00616 | 96.1 |
| 4BM1_Chr1_00600 | Two component transcriptional regulator | 4F5_Chr1_00617 | 93.3 |
| 4BM1_Chr1_00601 | Integral membrane sensor signal transduction histidine kinase | 4F5_Chr1_00618 | 93 |
| 4BM1_Chr1_00602 | Hypothetical protein | 4F5_Chr1_00619 | 88.9 |
| 4BM1_Chr1_00603 | ABC transporter substrate-binding protein | 4F5_Chr1_00620 | 93.6 |
| 4BM1_Chr1_00604 | glpT; glycerol-3-phosphate transporter | 4F5_Chr1_00621 | 89.6 |
表3 4BM1菌株胞外多糖生物合成基因簇的预测
Table 3 Prediction of gene cluster for exopolysaccharide synthesis in 4BM1 strain
| 基因编号 Gene ID | 功能预测 Predicted function | 4F5菌株中预测的胞外多糖基因簇 Predicted eps gene cluster in Bt 4F5 strain | 4BM1与4F5之间的氨基酸序列相似性Amino acid identity between 4BM1 and 4F5/% |
|---|---|---|---|
| 4BM1_Chr1_00581 | EpsC | 4F5_Chr1_00600 | N/A |
| 4BM1_Chr1_00582 | UTP-glucose-1-phosphate uridylyltransferase(EC:2.7.7.9) | 4F5_Chr1_00601 | 94.5 |
| 4BM1_Chr1_00583 | Lipopolysaccharide synthesis sugar transferase | 4F5_Chr1_00602 | 94.9 |
| 4BM1_Chr1_00584 | Nucleotide sugar dehydrogenase | 4F5_Chr1_00603 | 23 |
| 4BM1_Chr1_00585 | Asparagine synthase | 4F5_Chr1_00604 | N/A |
| 4BM1_Chr1_00586 | Polysaccharide biosynthesis protein | 4F5_Chr1_00605 | N/A |
| 4BM1_Chr1_00587 | Glycosyltransferase | 4F5_Chr1_00606 | N/A |
| 4BM1_Chr1_00588 | Hypothetical protein | 4F5_Chr1_00607 | N/A |
| 4BM1_Chr1_00589 | Hypothetical protein | 4F5_Chr1_00608 | N/A |
| 4BM1_Chr1_00590 | Glycosyltransferases group 1 | 4F5_Chr1_00609 | N/A |
| 4BM1_Chr1_00591 | Glycosyltransferase, group 1 family protein | 4F5_Chr1_00610 | N/A |
| 4BM1_Chr1_00592 | Glycosyltransferase | 4F5_Chr1_00611 | N/A |
| 4BM1_Chr1_00593 | Glucose-1-phosphate thymidylyltransferase(EC:2.7.7.24) | 4F5_Chr1_00612 | N/A |
| 4BM1_Chr1_00594 | dTDP-4-dehydrorhamnose 3,5-epimerase(EC:5.1.3.13) | None | |
| 4BM1_Chr1_00595 | dTDP-glucose 4,6-dehydratase | None | |
| 4BM1_Chr1_00596 | dTDP-4-dehydrorhamnose reductase(EC:1.1.1.133) | None | |
| 4BM1_Chr1_00597 | Membrane-bound transcriptional regulator LytR | 4F5_Chr1_00613 | 88.5 |
| 4BM1_Chr1_00598 | EPSX protein | 4F5_Chr1_00614 | 89.9 |
| 4BM1_Chr1_00599 | UDP-glucose 4-epimerase GalE | 4F5_Chr1_00616 | 96.1 |
| 4BM1_Chr1_00600 | Two component transcriptional regulator | 4F5_Chr1_00617 | 93.3 |
| 4BM1_Chr1_00601 | Integral membrane sensor signal transduction histidine kinase | 4F5_Chr1_00618 | 93 |
| 4BM1_Chr1_00602 | Hypothetical protein | 4F5_Chr1_00619 | 88.9 |
| 4BM1_Chr1_00603 | ABC transporter substrate-binding protein | 4F5_Chr1_00620 | 93.6 |
| 4BM1_Chr1_00604 | glpT; glycerol-3-phosphate transporter | 4F5_Chr1_00621 | 89.6 |
| 类型Type | 数量Amount |
|---|---|
| NPRS(non-ribosomal peptide synthetase cluster) | 4 |
| NPRS-like | 1 |
| Lasso peptide | 1 |
| Terpene | 1 |
| Ripp-like(Other unspecified ribosomally synthesised and post-translationally modified peptide product) | 2 |
| Beta-lactone(Beta-lactone containing protease inhibitor) | 1 |
| NI-siderophore(NRPS-independent, IucA/IucC-like siderophores) | 1 |
| LAP(Linear azol(in)e-containing peptides) | 1 |
| RRE-containing(RRE-element containing cluster) | 1 |
| Total | 13 |
表4 4BM1菌株次级代谢产物合成基因簇的预测
Table 4 Prediction of gene clusters for secondary metabolite biosynthesis of 4BM1 strain
| 类型Type | 数量Amount |
|---|---|
| NPRS(non-ribosomal peptide synthetase cluster) | 4 |
| NPRS-like | 1 |
| Lasso peptide | 1 |
| Terpene | 1 |
| Ripp-like(Other unspecified ribosomally synthesised and post-translationally modified peptide product) | 2 |
| Beta-lactone(Beta-lactone containing protease inhibitor) | 1 |
| NI-siderophore(NRPS-independent, IucA/IucC-like siderophores) | 1 |
| LAP(Linear azol(in)e-containing peptides) | 1 |
| RRE-containing(RRE-element containing cluster) | 1 |
| Total | 13 |
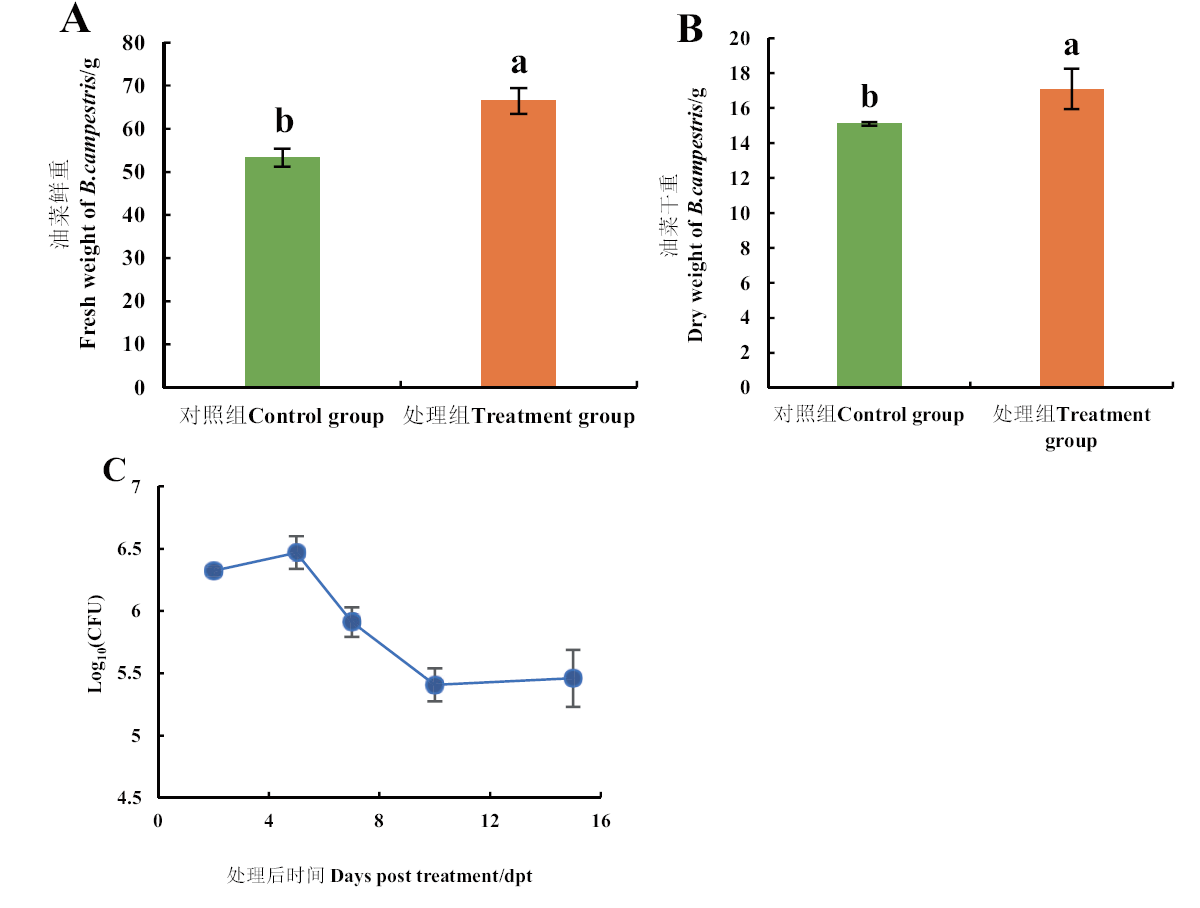
图3 4BM1菌株促生定殖能力的分析 A:4BM1菌株对油菜鲜重的影响;B:4BM1菌株对油菜干重的影响;C:4BM1菌株的定殖能力。所有数据为平均值+标准误(SE);不同小写字母表示在P<0.05水平上差异显著
Fig. 3 Analysis of growth-promoting and colonizing ability of 4BM1 strain A: Effect of 4BM1 strain on the fresh weight of B. campestris seedlings. B: Effect of 4BM1 strain on the dry weight of B. campestris seedlings. C: Ability of colonization of 4BM1 strain. All data are in the mean ± SE. Different lowercase letters indicate the difference at the level of P<0.05
| [1] | Bravo A, Likitvivatanavong S, Gill SS, et al. Bacillus thuringiensis: a story of a successful bioinsecticide[J]. Insect Biochem Mol Biol, 2011, 41(7): 423-431. |
| [2] | 王宇航, 束长龙, 耿丽丽, 等. 苏云金芽胞杆菌G033A产业化现状及应用前景分析[J]. 中国生物防治学报, 2020, 36(6): 837-841. |
| Wang YH, Shu CL, Geng LL, et al. Commercialization status and prospect analysis of Bacillus thuringiensis G033A product[J]. Chin J Biol Contr, 2020, 36(6): 837-841. | |
| [3] | Moazamian E, Bahador N, Azarpira N, et al. Anti-cancer parasporin toxins of new Bacillus thuringiensis against human colon(HCT-116)and blood(CCRF-CEM)cancer cell lines[J]. Curr Microbiol, 2018, 75(8): 1090-1098. |
| [4] | Cherif-Silini H, Silini A, Yahiaoui B, et al. Phylogenetic and plant-growth-promoting characteristics of Bacillus isolated from the wheat rhizosphere[J]. Ann Microbiol, 2016, 66(3): 1087-1097. |
| [5] | Nayak PS, Arakha M, Kumar A, et al. An approach towards continuous production of silver nanoparticles using Bacillus thuringiensis[J]. RSC Adv, 2016, 6(10): 8232-8242. |
| [6] | Bhatt P, Rene ER, Huang YH, et al. Indigenous bacterial consortium-mediated cypermethrin degradation in the presence of organic amendments and Zea mays plants[J]. Environ Res, 2022, 212(Pt A): 113137. |
| [7] | Martínez-Zavala SA, Barboza-Pérez UE, Hernández-Guzmán G, et al. Chitinases of Bacillus thuringiensis: phylogeny, modular structure, and applied potentials[J]. Front Microbiol, 2020, 10: 3032. |
| [8] | de la Fuente-Salcido NM, Casados-Vázquez LE, Barboza-Corona JE. Bacteriocins of Bacillus thuringiensis can expand the potential of this bacterium to other areas rather than limit its use only as microbial insecticide[J]. Can J Microbiol, 2013, 59(8): 515-522. |
| [9] | Roy A, Mahata D, Paul D, et al. Purification, biochemical characterization and self-assembled structure of a fengycin-like antifungal peptide from Bacillus thuringiensis strain SM1[J]. Front Microbiol, 2013, 4: 332. |
| [10] | Akram W, Mahboob A, Javed AA. Bacillus thuringiensis strain 199 can induce systemic resistance in tomato against Fusarium wilt[J]. Eur J Microbiol Immunol, 2013, 3(4): 275-280. |
| [11] | Wang ML, Geng LL, Jiao SM, et al. Bacillus thuringiensis exopolysaccharides induced systemic resistance against Sclerotinia sclerotiorum in Brassica campestris L[J]. Biol Contr, 2023, 183: 105267. |
| [12] | Wang ML, Geng LL, Sun XX, et al. Screening of Bacillus thuringiensis strains to identify new potential biocontrol agents against Sclerotinia sclerotiorum and Plutella xylostella in Brassica campestris L[J]. Biol Contr, 2020, 145: 104262. |
| [13] | Vlot AC, Sales JH, Lenk M, et al. Systemic propagation of immunity in plants[J]. New Phytol, 2021, 229(3): 1234-1250. |
| [14] | Hyakumachi M, Nishimura M, Arakawa T, et al. Bacillus thuringiensis suppresses bacterial wilt disease caused by Ralstonia solanacearum with systemic induction of defense-related gene expression in tomato[J]. Microbes Environ, 2013, 28(1): 128-134. |
| [15] | Wu LM, Huang ZY, Li X, et al. Stomatal closure and SA-, JA/ET-signaling pathways are essential for Bacillus amyloliquefaciens FZB42 to restrict leaf disease caused by Phytophthora nicotianae in Nicotiana benthamiana[J]. Front Microbiol, 2018, 9: 847. |
| [16] | Jiang CH, Fan ZH, Xie P, et al. Bacillus cereus AR156 extracellular polysaccharides served as a novel micro-associated molecular pattern to induced systemic immunity to pst DC3000 in Arabidopsis[J]. Front Microbiol, 2016, 7: 664. |
| [17] | Niu DD, Liu HX, Jiang CH, et al. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways[J]. Mol Plant Microbe Interact, 2011, 24(5): 533-542. |
| [18] | Takahashi H, Nakaho K, Ishihara T, et al. Transcriptional profile of tomato roots exhibiting Bacillus thuringiensis-induced resistance to Ralstonia solanacearum[J]. Plant Cell Rep, 2014, 33(1): 99-110. |
| [19] | Purdy LH. Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact[J]. Phytopathology, 1979, 69(8): 875. |
| [20] | Hossain MM, Sultana F, Li WQ, et al. Sclerotinia sclerotiorum(lib.) de bary: insights into the pathogenomic features of a global pathogen[J]. Cells, 2023, 12(7): 1063. |
| [21] | Benigni M, Bompeix G. Chemical and biological control of Sclerotinia sclerotiorum in witloof chicory culture[J]. Pest Manag Sci, 2010, 66(12): 1332-1336. |
| [22] | Silva LG, Camargo RC, Mascarin GM, et al. Dual functionality of Trichoderma: Biocontrol of Sclerotinia sclerotiorum and biostimulant of cotton plants[J]. Front Plant Sci, 2022, 13: 983127. |
| [23] | Zhao HZ, Zhou T, Xie JT, et al. Mycoparasitism illuminated by genome and transcriptome sequencing of Coniothyrium minitans, an important biocontrol fungus of the plant pathogen Sclerotinia sclerotiorum[J]. Microb Genom, 2020, 6(3): e000345. |
| [24] | Liu JF, Hu XW, He HL, et al. Digital gene expression profiling of the transcriptional response to Sclerotinia sclerotiorum and its antagonistic bacterium Bacillus amyloliquefaciens in soybean[J]. Front Microbiol, 2022, 13: 1025771. |
| [25] | Li H, Li HB, Bai Y, et al. The use of Pseudomonas fluorescens P13 to control Sclerotinia stem rot(Sclerotinia sclerotiorum)of oilseed rape[J]. J Microbiol, 2011, 49(6): 884-889. |
| [26] | Takahashi A, Casais C, Ichimura K, et al. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis[J]. Proc Natl Acad Sci USA, 2003, 100(20): 11777-11782. |
| [27] | Wang Z, Tan XL, Zhang ZY, et al. Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling[J]. Plant Sci, 2012, 184: 75-82. |
| [28] | Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions[J]. Genome Biol, 2013, 14(4): R36. |
| [29] | Florea L, Song L, Salzberg SL. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues[J]. F1000Research, 2013, 2: 188. |
| [30] | Anders S, Huber W. Differential expression analysis for sequence count data[J]. Genome Biol, 2010, 11(10): R106. |
| [31] | Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data[J]. Nat Methods, 2013, 10(6): 563-569. |
| [32] | Hyatt D, Chen GL, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification[J]. BMC Bioinformatics, 2010, 11: 119. |
| [33] | Emanuelsson O, Brunak S, von Heijne G, et al. Locating proteins in the cell using TargetP, SignalP and related tools[J]. Nat Protoc, 2007, 2(4): 953-971. |
| [34] | Lagesen K, Hallin P, Rødland EA, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes[J]. Nucleic Acids Res, 2007, 35(9): 3100-3108. |
| [35] | Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches[J]. Bioinformatics, 2013, 29(22): 2933-2935. |
| [36] | Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment[J]. Nucleic Acids Res, 2008, 36(Data-base issue): D480-D484. |
| [37] | Tatusov RL, Galperin MY, Natale DA, et al. The COG database: a tool for genome-scale analysis of protein functions and evolution[J]. Nucleic Acids Res, 2000, 28(1): 33-36. |
| [38] | Blin K, Medema MH, Kazempour D, et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers[J]. Nucleic Acids Res, 2013, 41(Web Server issue): W204-W212. |
| [39] | 王美玲, 耿丽丽, 段江燕, 等. 拮抗核盘菌Bt菌株的筛选及抑菌活性研究[J]. 植物保护, 2017, 43(6): 62-67. |
| Wang ML, Geng LL, Duan JY, et al. Screening of and studying on Bt strains with antagonistic activity against Sclerotinia sclerotiorum[J]. Plant Prot, 2017, 43(6): 62-67. | |
| [40] | Divi UK, Rahman T, Krishna P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways[J]. BMC Plant Biol, 2010, 10: 151. |
| [41] | Krishna P. Brassinosteroid-mediated stress responses[J]. J Plant Growth Regul, 2003, 22(4): 289-297. |
| [42] | Choi DW, Jung J, Ha YI, et al. Analysis of transcripts in methyl jasmonate-treated ginseng hairy roots to identify genes involved in the biosynthesis of ginsenosides and other secondary metabolites[J]. Plant Cell Rep, 2005, 23(8): 557-566. |
| [43] | Laranjeira S, Amorim-Silva V, Esteban A, et al. Arabidopsis squalene epoxidase 3(SQE3)complements SQE1 and is important for embryo development and bulk squalene epoxidase activity[J]. Mol Plant, 2015, 8(7): 1090-1102. |
| [44] | 翁启勇, 李开本. 诱导植物系统抗性研究及进展[J]. 福建农业学报, 1998, 13(4): 24-29. |
| Weng QY, Li KB. Study and progress in induction of systemic resistance of plant[J]. Fujian J Agric Sci, 1998, 13(4): 24-29. | |
| [45] | Wang K, Shu CL, Bravo A, et al. Development of an online genome sequence comparison resource for Bacillus cereus sensu lato strains using the efficient composition vector method[J]. Toxins, 2023, 15(6): 393. |
| [1] | 王芳, 于璐, 齐泽铮, 周长军, 于吉东. 大豆镰刀菌根腐病拮抗菌的筛选及生防效果[J]. 生物技术通报, 2024, 40(7): 216-225. |
| [2] | 范宗强, 冯靖涵, 郑丽雪, 王硕, 彭向前, 陈芳. 枯草芽孢杆菌B579对黄瓜枯萎病的防治及其诱导抗性研究[J]. 生物技术通报, 2024, 40(7): 226-234. |
| [3] | 徐伟芳, 李贺宇, 张慧, 何仔昂, 高文恒, 谢紫洋, 王传文, 尹登科. 生防细菌HX0037对栝楼炭疽病的防病能力及其机制[J]. 生物技术通报, 2024, 40(4): 228-241. |
| [4] | 许沛冬, 易剑锋, 陈迪, 潘磊, 谢丙炎, 赵文军. 贝莱斯芽孢杆菌生防次级代谢产物研究进展[J]. 生物技术通报, 2024, 40(3): 75-88. |
| [5] | 马云涛, 胡丽娜, 孙文婧, 唐莲庚, 孙思远, 邓欣雨, 孙黎. 梨火疫病拮抗菌JK2的筛选鉴定及发酵条件优化[J]. 生物技术通报, 2024, 40(11): 202-213. |
| [6] | 叶柳健, 贺愉岚, 王小虎, 韦圣博, 何双, 朱绮霞, 卢洁, 周礼芹. 解淀粉芽孢杆菌YK3对沃柑溃疡病的防效及叶际细菌群落相关性的影响[J]. 生物技术通报, 2024, 40(11): 248-258. |
| [7] | 李希, 边子俊, 宁周神, 刘红雨, 曾槟, 董伟. 离子型稀土矿根际芽孢杆菌的促生作用研究[J]. 生物技术通报, 2024, 40(11): 259-268. |
| [8] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [9] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [10] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [11] | 章乐乐, 王冠, 柳凤, 胡汉桥, 任磊. 芒果炭疽病拮抗菌分离、鉴定及生防机制研究[J]. 生物技术通报, 2023, 39(4): 277-287. |
| [12] | 易希, 廖红东, 郑井元. 植物内生真菌防治根结线虫研究进展[J]. 生物技术通报, 2023, 39(3): 43-51. |
| [13] | 王伟宸, 赵进, 黄薇颐, 郭芯竹, 李婉颖, 张卓. 芽胞杆菌代谢产物防治三种常见植物病原真菌的研究进展[J]. 生物技术通报, 2023, 39(3): 59-68. |
| [14] | 杨东亚, 祁瑞雪, 李昭轩, 林薇, 马慧, 张雪艳. 黄瓜茄病镰刀菌拮抗芽孢杆菌的筛选、鉴定及促生效果[J]. 生物技术通报, 2023, 39(2): 211-220. |
| [15] | 罗宁, 焦阳, 茆振川, 李惠霞, 谢丙炎. 木霉菌对根结线虫和孢囊线虫防治机理研究进展[J]. 生物技术通报, 2023, 39(2): 35-50. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||