








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 270-281.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0050
收稿日期:2024-01-15
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
郭红光,男,博士,教授,研究方向:煤系资源微生物开发、矿井灾害生物防治、矿山生态修复;E-mail: guohg_tyut@163.com作者简介:刘丁瑞,女,硕士研究生,研究方向:微生物增产煤层气;E-mail: 1786013906@qq.com
基金资助:
LIU Ding-rui1,2( ), GUO Hong-guang1,2(
), GUO Hong-guang1,2( ), GONG Kai-yi1,2
), GONG Kai-yi1,2
Received:2024-01-15
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】芳香化合物代谢是微生物降解煤产甲烷的限制因素。为了提高煤的生物甲烷产量,经过富集、驯化获得煤降解产甲烷菌群(RI)和菲降解功能菌群,并通过二者配伍获得复配菌群(CM)。【方法】采用宏基因组与宏转录组相结合方法分析CM与RI的菌群结构及代谢途径的异同。【结果】复配后菌群的甲烷产量明显提高,增产114.55%。复配显著提高了芳香化合物降解菌的占比,如Pseudomonas的占比高达63.49%;同时提高了优势菌的代谢活性以及芳香化合物代谢途径中各关键酶的合成和表达。CM中芳香族化合物降解途径的基因丰度是RI的1.65倍,基因表达丰度是RI的6.34倍(P<0.05)。其中,关键酶EC:1.13.11.2基因丰度和表达丰度分别是RI的2.24、62倍。这些酶表达丰度的增加促使更多的芳香族化合物代谢为丙酮酸。复配同时增强了丙酮酸代谢为乙酰辅酶A过程的基因表达,该代谢途径中关键酶EC:1.2.4.1的表达丰度在CM中可达到RI的14.70倍。CM中各产甲烷途径的基因表达丰度也高于RI,是RI的2.66-7.10倍。【结论】复配富集了芳香化合物降解菌,并显著提高了芳香化合物降解产甲烷整个代谢途径中基因丰度,尤其是基因表达丰度,从而提高甲烷产量。
刘丁瑞, 郭红光, 弓凯仪. 复配菌群降解煤产甲烷的宏基因组与宏转录组分析[J]. 生物技术通报, 2024, 40(9): 270-281.
LIU Ding-rui, GUO Hong-guang, GONG Kai-yi. Metagenomic and Metatranscriptomic Analysis of Methanogenesis from Coal Degradation by Compounded Microflora[J]. Biotechnology Bulletin, 2024, 40(9): 270-281.
图2 RI和CM在域水平的分布图 A:RI和CM在宏基因域水平的分布图;B:RI和CM在宏转录域水平的分布图
Fig. 2 Distribution of RI and CM at domain level A: Distribution of RI and CM at the level of metagenomic domains; B: Distribution of RI and CM at the level of metatranscriptomic domains
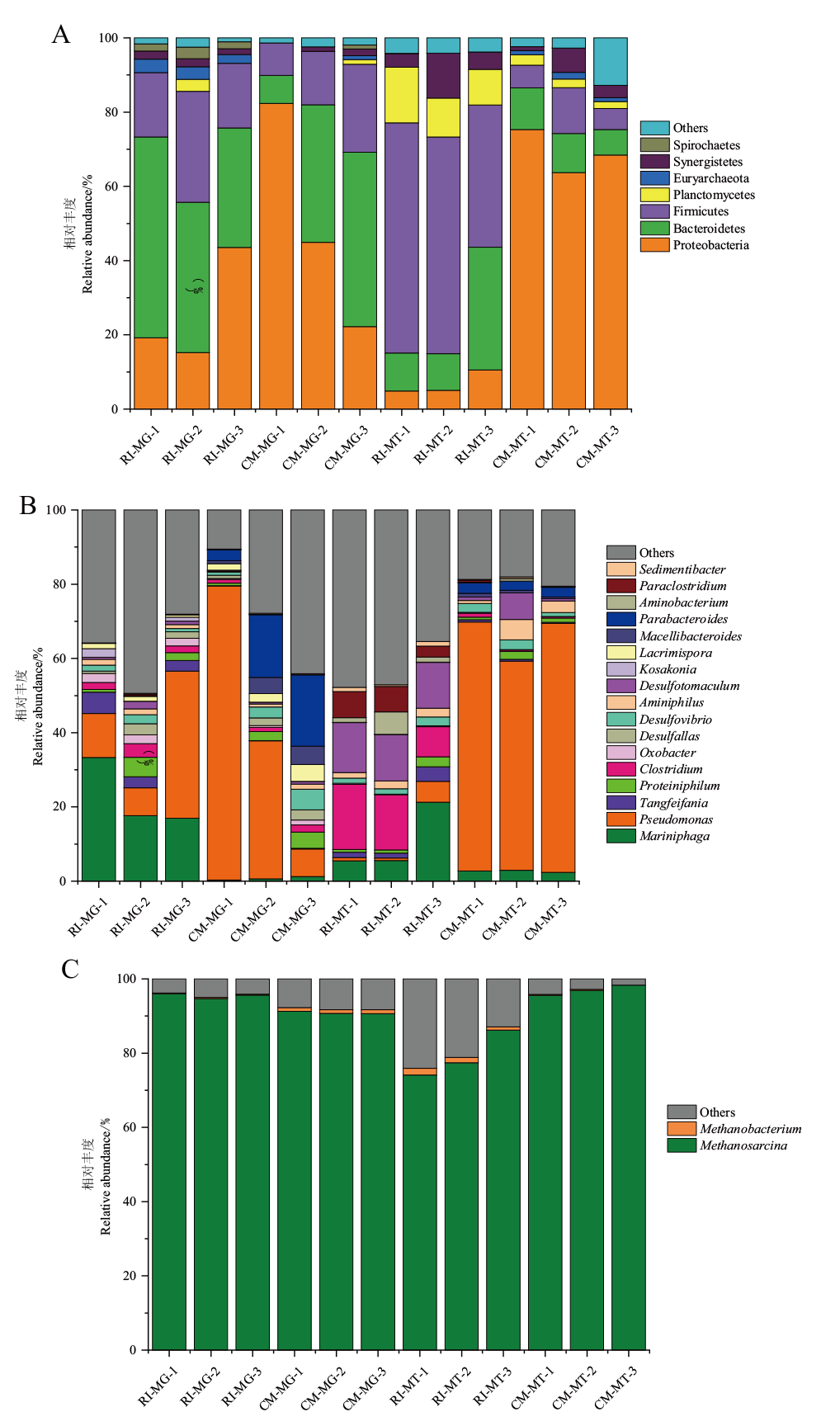
图3 RI和CM三组平行样在门水平(A)、细菌属水平(B)和古菌属水平(C)的微生物组成图 MG:宏基因组;MT:宏转录组,下同
Fig. 3 Microbial composition maps of three parallel groups of RI and CM samples at phylum(A), bacterial genus(B), and archaeal genus(C) MG: Metagenomic; MT: metatranscriptomic,the same below
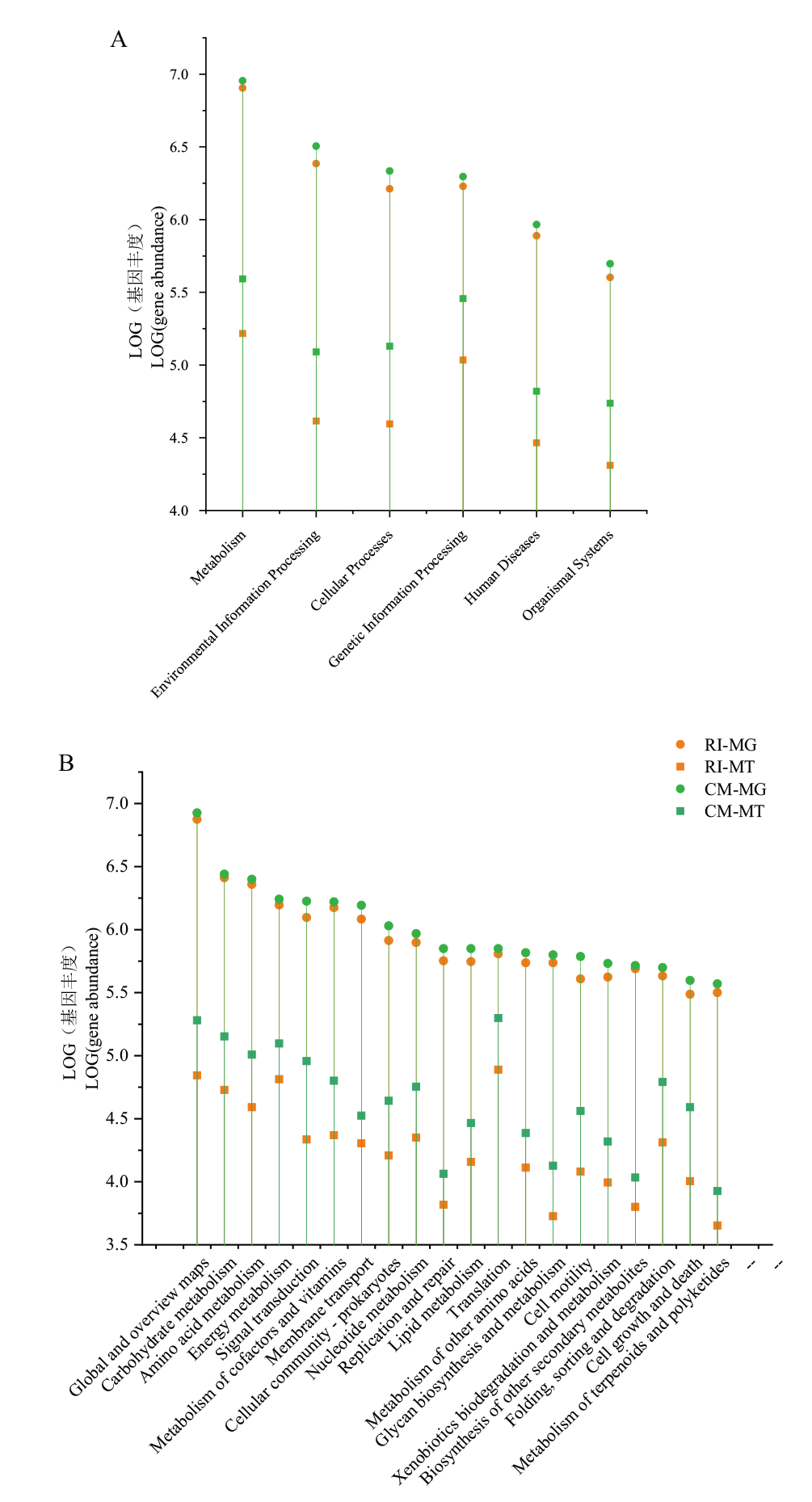
图4 生物反应器RI和CM中一级(A)和二级(B)代谢系统之间基因存在和表达绝对丰度 Y轴刻度代表每个类别的序列丰度(带注释的序列)的对数
Fig. 4 Absolute abundance of gene presence and expression between primary(A)and secondary(B)metabolic systems in bioreactor RI and CM The Y-axis scale refers to the logarithm of sequence abundance(sequences with annotations)for each category
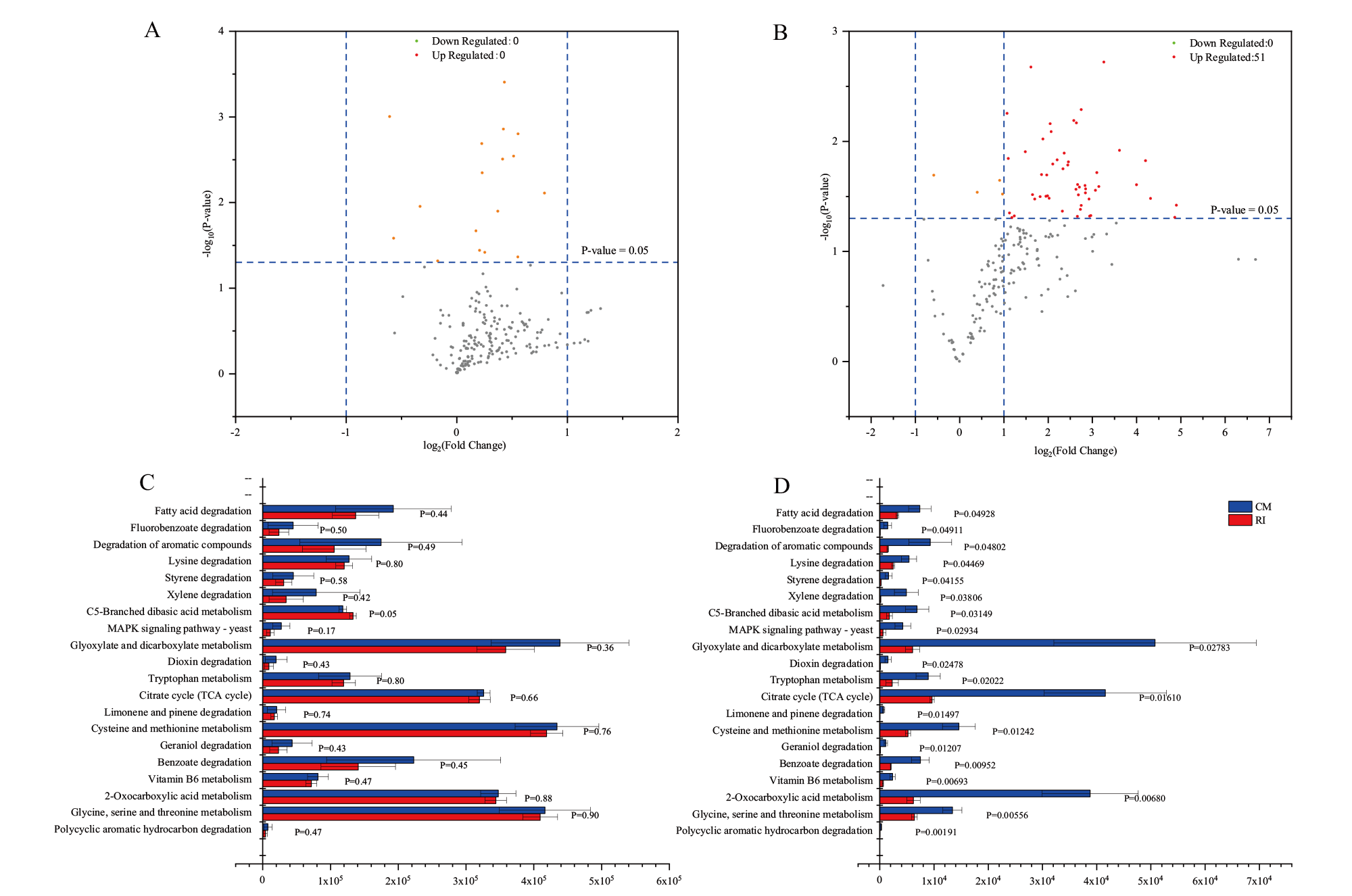
图5 三级通路上RI和CM代谢通路差异性分析 A:三级通路中宏基因KEGG火山图;B:三级通路中宏转录KEGG火山图;C:煤降解产甲烷代谢途径宏基因丰度比较;D:煤降解产甲烷代谢途径宏转录表达丰度比较
Fig. 5 Differential analysis of RI and CM metabolic pathways on tertiary pathways A:Volcano plot of KEGG in metagenomic in tertiary pathways. B: Volcano plot of KEGG in metatranscriptomic in tertiary pathways. C: Comparison of abundance of coal-degrading methane-producing metabolic pathways in metagenomic. D: Comparison of abundance of coal-degrading methane-producing metabolic pathways in metatranscriptomic

图6 芳香族化合物代谢途径及基因丰度和基因表达丰度图 柱状图代表每个基因丰度和表达丰度的对数,下同
Fig. 6 Aromatic compound metabolic pathways and plots of gene abundance and gene expression abundance The bar graphs refer to the logarithm of abundance and expression abundance for each gene, the same below
| 模块Module | RI-MG | RI-MT | CM-MG | CM-MT | 功能描述 Function |
|---|---|---|---|---|---|
| M00422 | 7578 | 1453.027 | 2252 | 6649.955 | Acetyl-CoA pathway, CO2 => acetyl-CoA |
| M00569 | 15455.33 | 114.273 | 34242 | 2434.298 | Catechol meta-cleavage, catechol => acetyl-CoA / 4-methylcatechol => propanoyl-CoA |
| M00036 | 79514 | 704.217 | 98381.33 | 1709.537 | Leucine degradation, leucine => acetoacetate + acetyl-CoA |
| M00013 | 10171.33 | 16.876 | 21789.33 | 957.577 | Malonate semialdehyde pathway, propanoyl-CoA => acetyl-CoA |
| M00032 | 32807.33 | 343.8167 | 37454.67 | 2953.905 | Lysine degradation, lysine => saccharopine => acetoacetyl-CoA |
| M00307 | 70726 | 1861.692 | 64192 | 3460.332 | Pyruvate oxidation, pyruvate => acetyl-CoA |
| M00086 | 42996 | 780.0583 | 50578 | 1389.26 | Beta-oxidation, acyl-CoA synthesis |
表1 乙酰辅酶A主要生成途径及途径基因丰度和基因的表达丰度
Table 1 Main production pathways of acetyl coenzyme A and abundance of genes and expression abundance of genes in the routes
| 模块Module | RI-MG | RI-MT | CM-MG | CM-MT | 功能描述 Function |
|---|---|---|---|---|---|
| M00422 | 7578 | 1453.027 | 2252 | 6649.955 | Acetyl-CoA pathway, CO2 => acetyl-CoA |
| M00569 | 15455.33 | 114.273 | 34242 | 2434.298 | Catechol meta-cleavage, catechol => acetyl-CoA / 4-methylcatechol => propanoyl-CoA |
| M00036 | 79514 | 704.217 | 98381.33 | 1709.537 | Leucine degradation, leucine => acetoacetate + acetyl-CoA |
| M00013 | 10171.33 | 16.876 | 21789.33 | 957.577 | Malonate semialdehyde pathway, propanoyl-CoA => acetyl-CoA |
| M00032 | 32807.33 | 343.8167 | 37454.67 | 2953.905 | Lysine degradation, lysine => saccharopine => acetoacetyl-CoA |
| M00307 | 70726 | 1861.692 | 64192 | 3460.332 | Pyruvate oxidation, pyruvate => acetyl-CoA |
| M00086 | 42996 | 780.0583 | 50578 | 1389.26 | Beta-oxidation, acyl-CoA synthesis |
| 样本名称Sample | M00357 | M00356 | M00567 | M00563 |
|---|---|---|---|---|
| RI-MG | 72573 | 48854 | 50116 | 46484 |
| RI-MT | 7123 | 1821 | 2040 | 1703 |
| CM-MG | 63086 | 28250 | 30324 | 28090 |
| CM-MT | 18937 | 12936 | 11077 | 10457 |
表2 产甲烷各个途径的基因含量及表达
Table 2 Gene content and expression of each pathway of methane production
| 样本名称Sample | M00357 | M00356 | M00567 | M00563 |
|---|---|---|---|---|
| RI-MG | 72573 | 48854 | 50116 | 46484 |
| RI-MT | 7123 | 1821 | 2040 | 1703 |
| CM-MG | 63086 | 28250 | 30324 | 28090 |
| CM-MT | 18937 | 12936 | 11077 | 10457 |
| [1] |
徐凤银, 侯伟, 熊先钺, 等. 中国煤层气产业现状与发展战略[J]. 石油勘探与开发, 2023, 50(4): 669-682.
doi: 10.11698/PED.20220856 |
| Xu FY, Hou W, Xiong XY, et al. The status and development strategy of coalbed methane industry in China[J]. Petrol Explor Dev, 2023, 50(4): 669-682. | |
| [2] | 范鹏鹏, 刘晓, 陈贞龙, 等. 中美煤层气勘探开发现状对比及启示[J]. 现代化工, 2023, 43(8): 22-25, 30. |
| Fan PP, Liu X, Chen ZL, et al. Comparison and enlightenment of current situation of coalbed methane exploration and development between China and the United States[J]. Mod Chem Ind, 2023, 43(8): 22-25, 30. | |
| [3] | 薛建英. 加强企业用地保障优化矿业权登记管理山西加大煤层气勘查开采支持力度[J]. 华北自然资源, 2023(5): 161. |
| Xue JY. Strengthen enterprise land security, optimize mining right registration management, and increase support for coalbed methane exploration and exploitation in Shanxi[J]. Huabei Nat Resour, 2023(5): 161. | |
| [4] | 苏现波, 夏大平, 赵伟仲, 等. 煤层气生物工程研究进展[J]. 煤炭科学技术, 2020, 48(6): 1-30. |
| Su XB, Xia DP, Zhao WZ, et al. Research advances of coalbed gas bioengineering[J]. Coal Sci Technol, 2020, 48(6): 1-30. | |
| [5] | Zhang Y, Zhang H, Li Y, et al. Study on increasing production of bio-methane from middle/high rank coal pretreated with hydrogen peroxide[J]. Coal Science and Technology, 2019, 47(09): 262-7. |
| [6] | Zhang J, Zhang H, Wang K, et al. Research progress of pretreatment technology for coal biotransformation[J]. Coal Technology, 2016, 35(11): 317-9. |
| [7] | Gong KY, Zhang YX, Guo HG, et al. Enhancing biomethane production from lignite by an anaerobic polycyclic aromatic hydrocarbon degrading fungal flora enriched from produced water[J]. Front Microbiol, 2022, 13: 899863. |
| [8] | Guo HG, Chen C, Liang WG, et al. Enhanced biomethane production from anthracite by application of an electric field[J]. Int J Coal Geol, 2020, 219: 103393. |
| [9] | Guo HG, Cheng YT, Huang ZX, et al. Factors affecting co-degradation of coal and straw to enhance biogenic coalbed methane[J]. Fuel, 2019, 244: 240-246. |
| [10] | Liu XS, Liu YJ, Tang H, et al. Microbial metabolism regulation on the efficient degradation of aromatic compounds for biochemical treatment process of coal chemical wastewater in pilot scale[J]. Environ Pollut, 2023, 331(Pt 2): 121872. |
| [11] | Ge XY, Ma XF, Wu ZS, et al. Modification of coal-based activated carbon with nitric acid using microwave radiation for adsorption of phenanthrene and naphthalene[J]. Res Chem Intermed, 2015, 41(10): 7327-7347. |
| [12] | Mai ZM, Wang L, Li QQ, et al. Biodegradation and metabolic pathway of phenanthrene by a newly isolated bacterium Gordonia sp. SCSIO19801[J]. Biochem Biophys Res Commun, 2021, 585: 42-47. |
| [13] | Avramova T, Sotirova A, Galabova D, et al. Effect of Triton X-100 and rhamnolipid PS-17 on the mineralization of phenanthrene by Pseudomonas sp. cells[J]. Int Biodeterior Biodegrad, 2008, 62(4): 415-420. |
| [14] | 张莉, 徐智敏, 孙亚军, 等. 鄂尔多斯典型煤矿不同功能区水化学与微生物群落特征及环境响应[J]. 煤炭科学技术, 2023, 51(12):180-196. |
| Zhang L, Xu ZM, Sun YJ, et al. Hydrochemistry and microbial community characteristics and environmental response in different functional zones of a typical coal mine in Ordos[J]. Coal Science and Technology, 2023, 51(12): 180-196. | |
| [15] | 陈家玉, 桂和荣, 郭艳, 等. 淮北煤田深层地下水微生物群落特征及其水源示踪意义[J]. 煤炭学报, 2023, 48(9):3503-12. |
| Chen JY, Gui HR, Guo Y, et al. Microbial community characteristics of deep groundwater in Huaibel coalfield and its significance in water source tracing[J]. Joumnal of China Coal Society, 2023, 48(9):3503-12. | |
| [16] |
Mukherjee A, Reddy MS. Metatranscriptomics: an approach for retrieving novel eukaryotic genes from polluted and related environments[J]. 3 Biotech, 2020, 10(2): 71.
doi: 10.1007/s13205-020-2057-1 pmid: 32030340 |
| [17] | Jha P, Ghosh S, Vidyarthi AS, et al. Unravelling the microbial community structure and function of coal-bed methane producing formation water of Jharia coal mines using metagenomics approach[J]. Fuel, 2022, 317: 123459. |
| [18] | Liu BJ, Wang YW, Li Y, et al. Improved formation of biogenic methane by cultivable bacteria in highly volatile bituminous coals[J]. J Clean Prod, 2022, 366: 132900. |
| [19] | Guo HY, Jia WQ, Chen ZH, et al. Analysis on methane production from various coal slime fermentations based on metagenomics[J]. J Environ Manage, 2023, 343: 118058. |
| [20] | Niu SY, Yang JY, McDermaid A, et al. Bioinformatics tools for quantitative and functional metagenome and metatranscriptome data analysis in microbes[J]. Brief Bioinform, 2018, 19(6): 1415-1429. |
| [21] | Chung YW, Gwak HJ, Moon S, et al. Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses[J]. PLoS One, 2020, 15(1): e0227886. |
| [22] | Kamke J, Kittelmann S, Soni P, et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation[J]. Microbiome, 2016, 4(1): 56. |
| [23] | Crovadore J, Soljan V, Calmin G, et al. Metatranscriptomic and metagenomic description of the bacterial nitrogen metabolism in waste water wet oxidation effluents[J]. Heliyon, 2017, 3(10): e00427. |
| [24] | Feng X, Zhang ZZ, Guo HG, et al. Enhancement of biogenic methane production from coal using supercritical CO2[J]. J Supercrit Fluids, 2023, 201: 106023. |
| [25] | Chen SF, Zhou YQ, Chen YR, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. |
| [26] |
Li DH, Liu CM, Luo RB, et al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph[J]. Bioinformatics, 2015, 31(10): 1674-1676.
doi: 10.1093/bioinformatics/btv033 pmid: 25609793 |
| [27] |
Fu LM, Niu BF, Zhu ZW, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152.
doi: 10.1093/bioinformatics/bts565 pmid: 23060610 |
| [28] |
Li RQ, Li YR, Kristiansen K, et al. SOAP: short oligonucleotide alignment program[J]. Bioinformatics, 2008, 24(5): 713-714.
doi: 10.1093/bioinformatics/btn025 pmid: 18227114 |
| [29] |
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND[J]. Nat Methods, 2015, 12(1): 59-60.
doi: 10.1038/nmeth.3176 pmid: 25402007 |
| [30] |
Wu JY, Gao WM, Johnson RH, et al. Integrated metagenomic and metatranscriptomic analyses of microbial communities in the meso- and bathypelagic realm of North Pacific Ocean[J]. Mar Drugs, 2013, 11(10): 3777-3801.
doi: 10.3390/md11103777 pmid: 24152557 |
| [31] | Yang G, Yin YN, Wang JL. Microbial community diversity during fermentative hydrogen production inoculating various pretreated cultures[J]. Int J Hydrog Energy, 2019, 44(26): 13147-13156. |
| [32] | Li JB, Meng DL, Wang XZ, et al. Sources and succession of microorganisms in industrial coal flotation system[J]. Fuel, 2023, 342: 127917. |
| [33] | Ruan MY, Hu ZQ, Zhu Q, et al. 16S rDNA sequencing-based insights into the bacterial community structure and function in co-existing soil and coal gangue[J]. Microorganisms, 2023, 11(9): 2151. |
| [34] | Wang DX, Han HJ, Han YX, et al. Enhanced treatment of Fischer-Tropsch(F-T)wastewater using the up-flow anaerobic sludge blanket coupled with bioelectrochemical system: effect of electric field[J]. Bioresour Technol, 2017, 232: 18-26. |
| [35] | Li WW, Khalid H, Zhu Z, et al. Methane production through anaerobic digestion: participation and digestion characteristics of cellulose, hemicellulose and lignin[J]. Appl Energy, 2018, 226: 1219-1228. |
| [36] |
Laothanachareon T, Kanchanasuta S, Mhuanthong W, et al. Analysis of microbial community adaptation in mesophilic hydrogen fermentation from food waste by tagged 16S rRNA gene pyrosequencing[J]. J Environ Manage, 2014, 144: 143-151.
doi: 10.1016/j.jenvman.2014.05.019 pmid: 24945701 |
| [37] | Aüllo T, Berlendis S, Lascourrèges JF, et al. New bio-indicators for long term natural attenuation of monoaromatic compounds in deep terrestrial aquifers[J]. Front Microbiol, 2016, 7: 122. |
| [38] | Orem WH, Tatu CA, Lerch HE, et al. Organic compounds in produced waters from coalbed natural gas wells in the Powder River Basin, Wyoming, USA[J]. Appl Geochem, 2007, 22(10): 2240-2256. |
| [39] | Ulrich G, Bower S. Active methanogenesis and acetate utilization in Powder River Basin coals, United States[J]. Int J Coal Geol, 2008, 76(1/2): 25-33. |
| [40] | Zhu JG, Liu RY, Cao N, et al. Mycobacterial metabolic characteristics in a water meter biofilm revealed by metagenomics and metatranscriptomics[J]. Water Res, 2019, 153: 315-323. |
| [41] | Zhao HP, Liang SH, Yang XE. Isolation and characterization of catechol 2, 3-dioxygenase genes from phenanthrene degraders Sphingomonas sp. ZP1 and Pseudomonas sp. ZP2[J]. Environ Technol, 2011, 33(15-16): 1895-1901. |
| [42] | Grifoll M, Selifonov SA, Chapman PJ. Evidence for a novel pathway in the degradation of fluorene by Pseudomonas sp. strain F274[J]. Appl Environ Microbiol, 1994, 60(7): 2438-2449. |
| [43] | Lin Q, De Vrieze J, He GH, et al. Temperature regulates methane production through the function centralization of microbial community in anaerobic digestion[J]. Bioresour Technol, 2016, 216: 150-158. |
| [44] | Liang JS, Nabi M, Zhang PY, et al. Promising biological conversion of lignocellulosic biomass to renewable energy with rumen microorganisms: a comprehensive review[J]. Renew Sustain Energy Rev, 2020, 134: 110335. |
| [45] | Zhong YJ, He JG, Wu F, et al. Metagenomic analysis reveals the size effect of magnetite on anaerobic digestion of waste activated sludge after thermal hydrolysis pretreatment[J]. Sci Total Environ, 2022, 851(Pt 1): 158133. |
| [46] | Chen Q, Liu CQ, Liu XY, et al. Magnetite enhances anaerobic digestion of high salinity organic wastewater[J]. Environ Res, 2020, 189: 109884. |
| [1] | 张迪, 鞠睿, 李丽梅, 王煜倩, 陈瑞, 王新一. 基于转录因子生物传感器在环境分析中的应用[J]. 生物技术通报, 2024, 40(6): 114-125. |
| [2] | 高云云, 杨海飞, 吕虎杰, 刘永鑫. 微生物组分析方法与功能挖掘[J]. 生物技术通报, 2024, 40(10): 98-107. |
| [3] | 张岩峰, 丁燕玲, 马应, 周小南, 杨朝云, 史远刚, 康晓龙. 肉牛剩余采食量相关瘤胃及粪便微生物特征比较分析[J]. 生物技术通报, 2023, 39(1): 295-304. |
| [4] | 鲁兆祥, 王夕冉, 连新磊, 廖晓萍, 刘雅红, 孙坚. 基于功能宏基因组学挖掘抗生素耐药基因研究进展[J]. 生物技术通报, 2022, 38(9): 17-27. |
| [5] | 张雨函, 范熠, 李婷婷, 庞爽, 刘为, 白可喻, 张西美. 基于宏基因组测序的植物叶表微生物富集及DNA提取方法[J]. 生物技术通报, 2022, 38(3): 256-263. |
| [6] | 马涛, 陆唯, 李松励, 樊霞. 畜禽微生物耐药组研究进展[J]. 生物技术通报, 2021, 37(1): 113-122. |
| [7] | 汪盼盼, 杨野, 刘迪秋, 崔秀明, 刘源. 宏基因组学在植物病害研究中的应用[J]. 生物技术通报, 2020, 36(12): 146-154. |
| [8] | 郭璟, 谢占玲, 罗涛, 薛治峰, 郭建娟, 李发雄, 张秀娟. 黄绿卷毛菇生境中矮嵩草内生真菌多样性比较研究[J]. 生物技术通报, 2019, 35(11): 109-117. |
| [9] | 陆洪省, 张雪, 高宇婷, 孙珮铭, 邱萌萌. 哈茨木霉SKD-ZX-1的鉴定、发酵及其生防效果[J]. 生物技术通报, 2019, 35(11): 132-140. |
| [10] | 王叶, 贾振华, 宋水山. 宏基因组学结合合成生物学法挖掘新型生物催化剂的研究进展[J]. 生物技术通报, 2018, 34(8): 35-42. |
| [11] | 李沛翰, 李鹏, 宋宏彬. 宏基因组学在传染病防控中的应用进展[J]. 生物技术通报, 2018, 34(3): 43-52. |
| [12] | 王朱珺, 王尚, 刘洋荧, 冯凯, 邓晔. 宏基因组技术在氮循环功能微生物分子检测研究中的应用[J]. 生物技术通报, 2018, 34(1): 1-14. |
| [13] | 宋伟凤, 李明聪, 高峥. 环境中微生物原位检测方法研究进展[J]. 生物技术通报, 2017, 33(10): 26-32. |
| [14] | 刘畅,罗著,张梦如,杨玉梅,刘娴,龚明,邹竹荣. 蓝藻橙色类胡萝卜素蛋白的基因克隆及在大肠杆菌中的异源表达和功能分析[J]. 生物技术通报, 2016, 32(7): 138-145. |
| [15] | 程福东, 丁啸, 李晟, 孙啸. 宏基因组样本数据的分析比较与分类[J]. 生物技术通报, 2016, 32(5): 1-10. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||