








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 243-252.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0373
韩乐乐1,2( ), 宋文迪1, 边嘉珅1, 李阳1, 杨双胜1, 陈紫怡1, 李晓薇1,2(
), 宋文迪1, 边嘉珅1, 李阳1, 杨双胜1, 陈紫怡1, 李晓薇1,2( )
)
收稿日期:2024-04-18
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
李晓薇,女,副教授,研究方向:植物抗逆基因工程;E-mail: xiaoweili1206@163.com作者简介:韩乐乐,男,硕士研究生,研究方向:植物抗逆基因工程;E-mail: 1491410856@qq.com
基金资助:
HAN Le-le1,2( ), SONG Wen-di1, BIAN Jia-shen1, LI Yang1, YANG Shuang-sheng1, CHEN Zi-yi1, LI Xiao-wei1,2(
), SONG Wen-di1, BIAN Jia-shen1, LI Yang1, YANG Shuang-sheng1, CHEN Zi-yi1, LI Xiao-wei1,2( )
)
Received:2024-04-18
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】大豆GmERD15c是ERD15转录因子家族成员之一,探究盐胁迫下转GmERD15c基因大豆株系的基因表达及物质代谢情况,揭示其与大豆耐盐性的关系。【方法】以转GmERD15c基因大豆株系及其受体大豆‘东农50’为材料,经盐胁迫处理后,采取根部进行转录组学和代谢组学分析。【结果】盐胁迫前后差异代谢物在类黄酮合成通路变化较大,胁迫后的代谢产物二氢山奈酚、儿茶素和槲皮素含量发生显著变化,且槲皮素含量在转GmERD15c基因株系中更高。同时,转录组学与代谢组学联合分析发现,GmERD15c基因与类黄酮生物合成相关酶关系密切。【结论】盐胁迫下,大豆GmERD15c基因可能通过调控类黄酮合成相关的酶类进而影响类黄酮的生物合成,而类黄酮可以减轻盐分引起的氧化应激,为后续揭示GmERD15c基因的耐盐机制提供了新线索。
韩乐乐, 宋文迪, 边嘉珅, 李阳, 杨双胜, 陈紫怡, 李晓薇. 转录组与代谢组联合分析揭示大豆GmERD15c参与盐胁迫下类黄酮的生物合成[J]. 生物技术通报, 2024, 40(10): 243-252.
HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome[J]. Biotechnology Bulletin, 2024, 40(10): 243-252.
| 代谢物级别 Metabolite level | 组别 Group | 代谢物数量 Number of metabolites | 上调数量 Up-regulated number | 下调数量 Down-regulated number | 总差异代谢物 Total differential metabolites |
|---|---|---|---|---|---|
| 一级差异代谢物 Primary differential metabolites | WT-mock VS OE-mock | 20 804 | 846 | 847 | 1 693 |
| WT-NaCl VS OE-NaCl | 20 804 | 840 | 735 | 1 575 | |
| 二级差异代谢物 Secondary differential metabolites | WT-mock VS OE-mock | 402 | 20 | 15 | 35 |
| WT-NaCl VS OE-NaCl | 402 | 31 | 21 | 52 |
表1 差异代谢物
Table 1 Differential metabolites
| 代谢物级别 Metabolite level | 组别 Group | 代谢物数量 Number of metabolites | 上调数量 Up-regulated number | 下调数量 Down-regulated number | 总差异代谢物 Total differential metabolites |
|---|---|---|---|---|---|
| 一级差异代谢物 Primary differential metabolites | WT-mock VS OE-mock | 20 804 | 846 | 847 | 1 693 |
| WT-NaCl VS OE-NaCl | 20 804 | 840 | 735 | 1 575 | |
| 二级差异代谢物 Secondary differential metabolites | WT-mock VS OE-mock | 402 | 20 | 15 | 35 |
| WT-NaCl VS OE-NaCl | 402 | 31 | 21 | 52 |

图3 转GmERD15c基因大豆类黄酮合成通路的代谢物变化 A:二级差异代谢物主要分类;B:转基因大豆代谢物KEGG分析;C:转基因大豆类黄酮化合物定量分析
Fig. 3 Metabolite variation in flavonoid synthesis pathway of soybean with transgenic GmERD15c gene A: Main classification of secondary differential metabolites. B: KEGG analysis of transgenic soybean metabolites. C: Quantitative analysis of transgenic soybean flavonoid compounds
| 对照组Control | 实验组Treat | 上调表达基因Up-regulated genes | 下调表达基因Down-regulated genes | 差异表达基因Total DEGs |
|---|---|---|---|---|
| WT-mock | OE-mock | 605 | 1 208 | 1 813 |
| WT-NaCl | OE-NaCl | 681 | 693 | 1 374 |
| OE-mock | OE-NaCl | 5 348 | 4 456 | 9 804 |
| WT-mock | WT-NaCl | 6 576 | 6 533 | 13 109 |
表2 大豆转录组差异基因分析
Table 2 Transcriptome differential gene analysis in soybean
| 对照组Control | 实验组Treat | 上调表达基因Up-regulated genes | 下调表达基因Down-regulated genes | 差异表达基因Total DEGs |
|---|---|---|---|---|
| WT-mock | OE-mock | 605 | 1 208 | 1 813 |
| WT-NaCl | OE-NaCl | 681 | 693 | 1 374 |
| OE-mock | OE-NaCl | 5 348 | 4 456 | 9 804 |
| WT-mock | WT-NaCl | 6 576 | 6 533 | 13 109 |

图4 大豆转录组差异基因Upset图 Number in each set表示每个比较组鉴定到的全部差异基因的数目。Number of each intersect表示多个比较组鉴定到的共有差异基因的数目,横坐标一个点表示该比较组鉴定到的特有差异基因的数目,横坐标多个点连线表示连线的多个比较组鉴定到的共有差异基因的数目。蓝色为1个组别,黄色为2个组别,红色为3个组别,绿色为4个组别
Fig. 4 Upset diagram of transcriptome differential genes in soybean Number in each set indicates the number of all differential genes identified in each comparison group. Number of each intersect indicates the number of common differences identified by multiple comparison groups, a point on the abscissa indicates the number of unique differences identified by the comparison group, and a line connecting multiple points on the abscissa indicates the number of common differences identified by multiple comparison groups connected by the line. Blue indicates one group, yellow indicates two groups, red indicates three groups, and green indicates four groups
| 通路ID Pathway ID | KEGG通路 KEGG pathway | 1级通路 Level 1 | 2级通路 Level 2 | 上调数量 Up_number | 下调数量 Down_number | 差异数量 DEG_number | 总数量 Total_number |
|---|---|---|---|---|---|---|---|
| gmx00910 | 氮代谢 | 新陈代谢 | 能量代谢 | 6 | 2 | 8 | 44 |
| gmx00260 | 甘氨酸、丝氨酸和苏氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 4 | 4 | 8 | 89 |
| gmx00940 | 苯丙烷类生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 8 | 7 | 15 | 260 |
| gmx00565 | 乙醚脂质代谢 | 新陈代谢 | 脂类代谢 | 5 | 0 | 5 | 43 |
| gmx00904 | 二萜类生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 1 | 4 | 5 | 45 |
| gmx00564 | 甘油磷脂代谢 | 新陈代谢 | 脂类代谢 | 9 | 1 | 10 | 164 |
| gmx00592 | 亚麻酸代谢 | 新陈代谢 | 脂类代谢 | 4 | 2 | 6 | 72 |
| gmx00908 | 玉米素生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 3 | 1 | 4 | 36 |
| gmx00350 | 酪氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 3 | 2 | 5 | 56 |
| gmx00270 | 半胱氨酸和蛋氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 6 | 3 | 9 | 154 |
| gmx00630 | 乙醛酸盐和二羧酸代谢 | 新陈代谢 | 碳水化合物代谢 | 0 | 6 | 6 | 87 |
| gmx00130 | 泛醌和其他萜类醌生物合成 | 新陈代谢 | 辅因子和维生素的代谢 | 4 | 1 | 5 | 68 |
| gmx00943 | 异黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 0 | 3 | 28 |
| gmx00941 | 类黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 2 | 5 | 77 |
表3 盐胁迫下转GmERD15c基因大豆转录组KEGG分析
Table 3 KEGG analysis of soybean transcriptome transgenic GmERD15c gene under salt stress
| 通路ID Pathway ID | KEGG通路 KEGG pathway | 1级通路 Level 1 | 2级通路 Level 2 | 上调数量 Up_number | 下调数量 Down_number | 差异数量 DEG_number | 总数量 Total_number |
|---|---|---|---|---|---|---|---|
| gmx00910 | 氮代谢 | 新陈代谢 | 能量代谢 | 6 | 2 | 8 | 44 |
| gmx00260 | 甘氨酸、丝氨酸和苏氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 4 | 4 | 8 | 89 |
| gmx00940 | 苯丙烷类生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 8 | 7 | 15 | 260 |
| gmx00565 | 乙醚脂质代谢 | 新陈代谢 | 脂类代谢 | 5 | 0 | 5 | 43 |
| gmx00904 | 二萜类生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 1 | 4 | 5 | 45 |
| gmx00564 | 甘油磷脂代谢 | 新陈代谢 | 脂类代谢 | 9 | 1 | 10 | 164 |
| gmx00592 | 亚麻酸代谢 | 新陈代谢 | 脂类代谢 | 4 | 2 | 6 | 72 |
| gmx00908 | 玉米素生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 3 | 1 | 4 | 36 |
| gmx00350 | 酪氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 3 | 2 | 5 | 56 |
| gmx00270 | 半胱氨酸和蛋氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 6 | 3 | 9 | 154 |
| gmx00630 | 乙醛酸盐和二羧酸代谢 | 新陈代谢 | 碳水化合物代谢 | 0 | 6 | 6 | 87 |
| gmx00130 | 泛醌和其他萜类醌生物合成 | 新陈代谢 | 辅因子和维生素的代谢 | 4 | 1 | 5 | 68 |
| gmx00943 | 异黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 0 | 3 | 28 |
| gmx00941 | 类黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 2 | 5 | 77 |
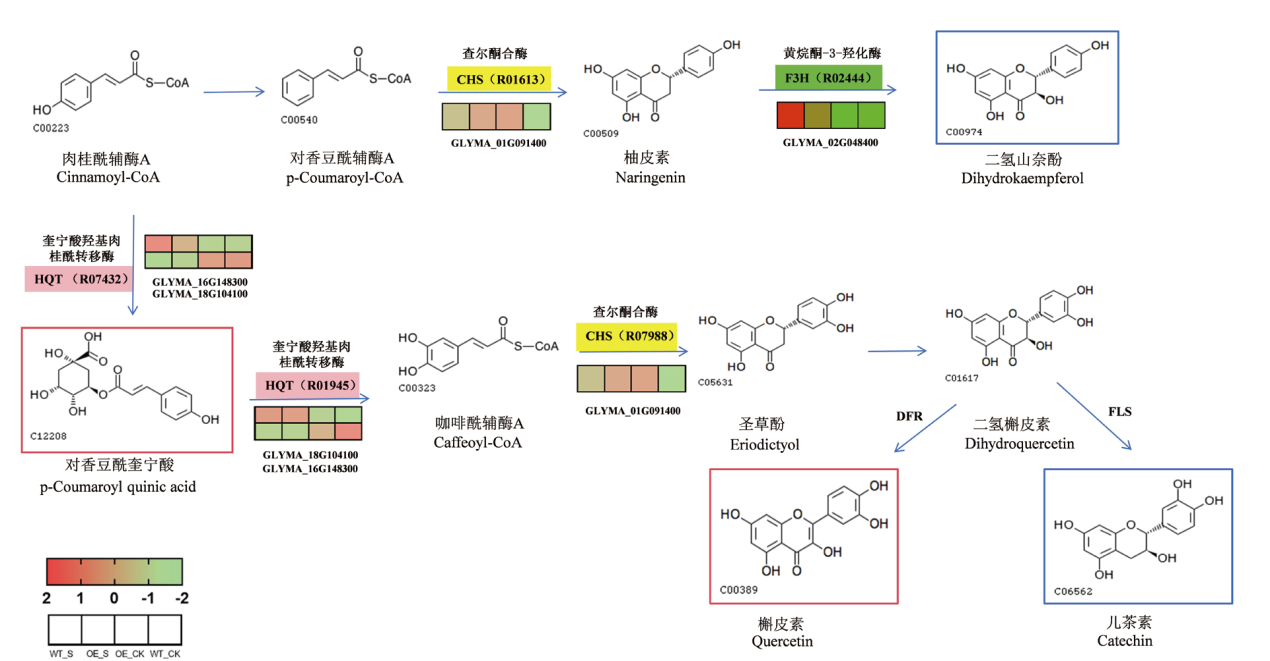
图5 转录代谢联合KEGG分析 底色表示mRNA,矩形表示代谢物。蓝色表示代谢物下调,红色表示代谢物上调,绿色表示mRNA下调,黄色mRNA上调,粉色表示多差异mRNA。括号中的编号表示酶的反应类型,R07432:奎宁酸羟基肉桂酰转移酶,R01945:奎宁酸酯O-(3,4-二羟基肉桂酰基)转移酶,R01613:4-香豆酰辅酶A丙二酰转移酶 R07988:咖啡酰辅酶A丙二酰转移酶 R02444:氧化还原酶
Fig. 5 Transcriptional metabolism combined with KEGG analysis The base color indicates mRNA and the rectangle indicates metabolites. Blue indicates metabolite downregulation, red indicates metabolite upregulation, green indicates mRNA downregulation, yellow indicates mRNA upregulation, and pink indicates polydifferential mRNA.The numbers in the parentheses indicate the types of enzyme reactions. R07432: Quinate hydroxycinnamoyltransferase.R01945: Quinate ester O-(3,4-dihydroxycinnamoyl)transferase. R01613: 4-coumaroyl-CoA:malate acyltransferase. R07988: Caffeoyl-CoA:malate acyltransferase. R02444: Oxidoreductase
| [1] | 王文月, 姚志鹏, 于洋, 等. 我国大豆种业科技创新发展现状及对策建议[J]. 中国农业科技导报, 2024, 26(3): 1-6. |
| Wang WY, Yao ZP, Yu Y, et al. Scientific and technological innovation of soybean seed industry in China: current situation and strategy[J]. J Agric Sci Technol, 2024, 26(3): 1-6. | |
| [2] |
Kumar A, Singh S, Gaurav AK, et al. Plant growth-promoting bacteria: biological tools for the mitigation of salinity stress in plants[J]. Front Microbiol, 2020, 11: 1216.
doi: 10.3389/fmicb.2020.01216 pmid: 32733391 |
| [3] |
Liang XY, Li JF, Yang YQ, et al. Designing salt stress-resilient crops: current progress and future challenges[J]. J Integr Plant Biol, 2024, 66(3): 303-329.
doi: 10.1111/jipb.13599 |
| [4] | Ahammed GJ, Yang YX. Anthocyanin-mediated arsenic tolerance in plants[J]. Environ Pollut, 2022, 292(Pt B): 118475. |
| [5] | Clayton WA, Albert NW, Thrimawithana AH, et al. UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants[J]. Plant J, 2018, 96(3): 503-517. |
| [6] | Yu WW, Liu HM, Luo JQ, et al. Partial root-zone simulated drought induces greater flavonoid accumulation than full root-zone simulated water deficiency in the leaves of Ginkgo biloba[J]. Environ Exp Bot, 2022, 201: 104998. |
| [7] | Li Q, Yu HM, Meng XF, et al. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis[J]. Plant Biol, 2018, 20(1): 10-19. |
| [8] | Li BZ, Fan RN, Guo SY, et al. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis[J]. Environ Exp Bot, 2019, 166: 103807. |
| [9] | Bian XH, Li W, Niu CF, et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis[J]. New Phytol, 2020, 225(1): 268-283. |
| [10] | Aalto MK, Helenius E, Kariola T, et al. ERD15—an attenuator of plant ABA responses and stomatal aperture[J]. Plant Sci, 2012, 182: 19-28. |
| [11] | Wu GF, Tian NF, She FW, et al. Characteristics analysis of Early Responsive to Dehydration genes in Arabidopsis thaliana(AtER-D)[J]. Plant Signal Behav, 2023, 18(1): 2105021. |
| [12] | Huang YM, Du BS, Yu MX, et al. Picea wilsonii NAC31 and DREB2A cooperatively activate ERD1 to modulate drought resistance in transgenic Arabidopsis[J]. Int J Mol Sci, 2024, 25(4): 2037. |
| [13] |
Kariola T, Brader G, Helenius E, et al. EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis[J]. Plant Physiol, 2006, 142(4): 1559-1573.
doi: 10.1104/pp.106.086223 pmid: 17056758 |
| [14] |
Jin T, Sun YY, Shan Z, et al. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean[J]. Plant Biotechnol J, 2021, 19(6): 1155-1169.
doi: 10.1111/pbi.13536 pmid: 33368860 |
| [15] | Slawinski L, Israel A, Artault C, et al. Responsiveness of early response to dehydration six-like transporter genes to water deficit in Arabidopsis thaliana leaves[J]. Front Plant Sci, 2021, 12: 708876. |
| [16] |
Farrant JM, Cooper K, Hilgart A, et al. A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa(Baker)[J]. Planta, 2015, 242(2): 407-426.
doi: 10.1007/s00425-015-2320-6 pmid: 25998524 |
| [17] | Ziaf K, Loukehaich R, Gong PJ, et al. A multiple stress-responsive gene ERD15 from Solanum pennellii confers stress tolerance in tobacco[J]. Plant Cell Physiol, 2011, 52(6): 1055-1067. |
| [18] | 曹沙沙. 大豆GmERD15c基因的克隆及其功能分析[D]. 长春: 吉林农业大学, 2022. |
| Cao SS. Cloning and functional analysis of GmERD15c gene in soybean[D]. Changchun: Jilin Agricultural University, 2022. | |
| [19] |
Wu JT, Lv SD, Zhao L, et al. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses[J]. Planta, 2023, 257(6): 108.
doi: 10.1007/s00425-023-04136-w pmid: 37133783 |
| [20] | Li SC. Novel insight into functions of ascorbate peroxidase in higher plants: more than a simple antioxidant enzyme[J]. Redox Biol, 2023, 64: 102789. |
| [21] | Speisky H, Shahidi F, Costa de Camargo A, et al. Revisiting the oxidation of flavonoids: loss, conservation or enhancement of their antioxidant properties[J]. Antioxidants, 2022, 11(1): 133. |
| [22] | Wang ZL, Wang S, Kuang Y, et al. A comprehensive review on phytochemistry, pharmacology, and flavonoid biosynthesis of Scutellaria baicalensis[J]. Pharm Biol, 2018, 56(1): 465-484. |
| [23] | Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions[J]. J Integr Plant Biol, 2021, 63(1): 180-209. |
| [24] | Uleberg E, Rohloff J, Jaakola L, et al. Effects of temperature and photoperiod on yield and chemical composition of northern and southern clones of bilberry(Vaccinium myrtillus L.)[J]. J Agric Food Chem, 2012, 60(42): 10406-10414. |
| [25] |
Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid[J]. Nat Biotechnol, 2004, 22(6): 746-754.
doi: 10.1038/nbt966 pmid: 15107863 |
| [26] | Guo J, Carrington Y, Alber A, et al. Molecular characterization of quinate and shikimate metabolism in Populus trichocarpa[J]. J Biol Chem, 2014, 289(34): 23846-23858. |
| [27] | Clé C, Hill LM, Niggeweg R, et al. Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance[J]. Phytochemistry, 2008, 69(11): 2149-2156. |
| [28] | Deng XB, Bashandy H, Ainasoja M, et al. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida[J]. New Phytol, 2014, 201(4): 1469-1483. |
| [29] | Zhang XB, Abrahan C, Colquhoun TA, et al. A proteolytic regulator controlling Chalcone synthase stability and flavonoid biosynthesis in Arabidopsis[J]. Plant Cell, 2017, 29(5): 1157-1174. |
| [30] |
Schijlen EGWM, de Vos CH, Martens S, et al. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits[J]. Plant Physiol, 2007, 144(3): 1520-1530.
doi: 10.1104/pp.107.100305 pmid: 17478633 |
| [31] | Song XY, Diao JJ, Ji J, et al. Molecular cloning and identification of a flavanone 3-hydroxylase gene from Lycium chinense, and its overexpression enhances drought stress in tobacco[J]. Plant Physiol Biochem, 2016, 98: 89-100. |
| [32] |
Tu YH, Liu F, Guo DD, et al. Molecular characterization of flavanone 3-hydroxylase gene and flavonoid accumulation in two chemotyped safflower lines in response to methyl jasmonate stimulation[J]. BMC Plant Biol, 2016, 16(1): 132.
doi: 10.1186/s12870-016-0813-5 pmid: 27286810 |
| [33] | Busche M, Acatay C, Martens S, et al. Functional characterisation of banana(Musa spp.) 2-oxoglutarate-dependent dioxygenases involved in flavonoid biosynthesis[J]. Front Plant Sci, 2021, 12: 701780. |
| [34] | Wang LX, Lui ACW, Lam PY, et al. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum[J]. Plant Biotechnol J, 2020, 18(11): 2170-2172. |
| [35] | Liu W, Feng Y, Yu S, et al. The flavonoid biosynthesis network in plants[J]. Int J Mol Sci, 2021, 22(23): 12824. |
| [36] | Mou JL, Zhang ZH, Qiu HJ, et al. Multiomics-based dissection of Citrus flavonoid metabolism using a Citrus reticulata × Poncirus trifoliata population[J]. Hortic Res, 2021, 8(1): 56. |
| [37] | Meng XY, Li YQ, Zhou TT, et al. Functional differentiation of duplicated flavonoid 3- O-glycosyltransferases in the flavonol and anthocyanin biosynthesis of Freesia hybrida[J]. Front Plant Sci, 2019, 10: 1330. |
| [38] | Singh P, Arif Y, Bajguz A, et al. The role of quercetin in plants[J]. Plant Physiol Biochem, 2021, 166: 10-19. |
| [39] |
Martens S, Preuss A, Matern U. Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L[J]. Phytochemistry, 2010, 71(10): 1040-1049.
doi: 10.1016/j.phytochem.2010.04.016 pmid: 20457455 |
| [40] | Yan HL, Pei XN, Zhang H, et al. MYB-mediated regulation of anthocyanin biosynthesis[J]. Int J Mol Sci, 2021, 22(6): 3103. |
| [41] | Hinojosa-Gómez J, San Martín-Hernández C, Heredia JB, et al. Anthocyanin induction by drought stress in the Calyx of Roselle cultivars[J]. Molecules, 2020, 25(7): 1555. |
| [42] | Chen WF, Xiao ZC, Wang YL, et al. Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus[J]. Hortic Res, 2021, 8(1): 173. |
| [43] |
Yan JH, Wang B, Zhong YP, et al. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis[J]. Plant Mol Biol, 2015, 89(1-2): 35-48.
doi: 10.1007/s11103-015-0349-3 pmid: 26231207 |
| [44] | Wang X, Dai WW, Liu C, et al. Evaluation of physiological coping strategies and quality substances in purple SweetPotato under different salinity levels[J]. Genes, 2022, 13(8): 1350. |
| [45] | Xu ZC, Wang M, Ren TT, et al. Comparative transcriptome analysis reveals the molecular mechanism of salt tolerance in Apocynum venetum[J]. Plant Physiol Biochem, 2021, 167: 816-830. |
| [1] | 王睿, 戚继. 整合组织学图像信息增强空间转录组细胞聚类的分辨率[J]. 生物技术通报, 2024, 40(8): 39-46. |
| [2] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| [3] | 高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149. |
| [4] | 王芳, 于璐, 齐泽铮, 周长军, 于吉东. 大豆镰刀菌根腐病拮抗菌的筛选及生防效果[J]. 生物技术通报, 2024, 40(7): 216-225. |
| [5] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [6] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [7] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [8] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [9] | 李灿, 蒋湘宁, 盖颖. 日本落叶松LkF3H2基因克隆及调控类黄酮代谢功能研究[J]. 生物技术通报, 2024, 40(2): 245-252. |
| [10] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| [11] | 姜宇舢, 兰倩, 王芳, 姜亮, 裴成成. 一个影响酪氨酸代谢藜麦突变体的鉴定[J]. 生物技术通报, 2024, 40(10): 253-261. |
| [12] | 何诗瑜, 曾仲大, 李博岩. 空间分辨代谢组学在疾病诊断研究中的应用进展[J]. 生物技术通报, 2024, 40(1): 145-159. |
| [13] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| [14] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [15] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||