








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 233-242.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0475
李欣格1,2( ), 王美霞2,3, 王晨阳2, 马桂根2,4, 周常勇3, 王亚南1(
), 王美霞2,3, 王晨阳2, 马桂根2,4, 周常勇3, 王亚南1( ), 周焕斌2,4,5(
), 周焕斌2,4,5( )
)
收稿日期:2024-05-21
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
王亚南,女,博士,教授,研究方向:植物病毒分子检测、果蔬病毒与寄主互作研究;E-mail: wyn3215347@163.com;作者简介:李欣格,女,硕士研究生,研究方向:基因组编辑技术开发与应用;E-mail: xingell@hotmail.com
基金资助:
LI Xin-ge1,2( ), WANG Mei-xia2,3, WANG Chen-yang2, MA Gui-gen2,4, ZHOU Chang-yong3, WANG Ya-nan1(
), WANG Mei-xia2,3, WANG Chen-yang2, MA Gui-gen2,4, ZHOU Chang-yong3, WANG Ya-nan1( ), ZHOU Huan-bin2,4,5(
), ZHOU Huan-bin2,4,5( )
)
Received:2024-05-21
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】在水稻中建立CRISPR/LanCas9 和 CRISPR/SLanCas9基因编辑系统,扩展CRISPR/Cas基因编辑工具箱。【方法】将来自动物乳酸杆菌KCTC 3501的LanCas9进行密码子优化,并且进一步通过蛋白融合重组LanCas9和SpCas9的活性结构域形成嵌合体SLanCas9,分别构建CRISPR/LanCas9和CRISPR/SLanCas9编辑工具;以OsWRKY45、OsCPK4、OsCPK6、OsCPK7、OsMPK8、OsGSK3、OsGSK4为靶基因,在NGG PAM或者NAG PAM设计20 nt或者24 nt的sgRNA,通过水稻遗传转化,检测分析其编辑效率。【结果】LanCas9在水稻中可以识别NGG PAM,结合20 nt的sgRNA编辑效率更高,对OsWRKY45基因的编辑效率为25.00%;融合的SLanCas9不仅能够识别NGG PAM,在NAG PAM位点也有一定的编辑效率,在不同PAM位点的编辑效率分别可达100%和39.58%。此外,SLanCas9还具有多重基因编辑能力,在NGG PAM位点的多重基因编辑效率高达74.07%。【结论】开发了具有自主知识产权的新型CRISPR/LanCas9和CRISPR/SLanCas9基因编辑技术。
李欣格, 王美霞, 王晨阳, 马桂根, 周常勇, 王亚南, 周焕斌. 基于CRISPR/LanCas9的水稻基因编辑系统的开发和优化[J]. 生物技术通报, 2024, 40(10): 233-242.
LI Xin-ge, WANG Mei-xia, WANG Chen-yang, MA Gui-gen, ZHOU Chang-yong, WANG Ya-nan, ZHOU Huan-bin. Development and Optimization of Genome Editing in Rice with CRISPR/LanCas9 System[J]. Biotechnology Bulletin, 2024, 40(10): 233-242.
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| OsMPK8-F1 | TGGCGGTGGAGGAGAGGTGA | Amplify the OsMPK8 target site in this research |
| OsMPK8-R1 | CAACCACCCATGACCATGAG | |
| OsWRKY45-F1 | TGCAGCCGCTCAAGGTGGAG | Amplify the OsWRKY45 target site in this research |
| OsWRKY45-R1 | TCAAACCCATAATGTCGTCC | |
| OsCPK6-F1 | AGAAACAGCATCTCTTGAGG | Amplify the OsCPK6 target site in this research |
| OsCPK6-R1 | GCTACTTGTTGCTGTGCTTG | |
| OsCPK4-F1 | TTCTCATCCCACACTGCGAC | Amplify two OsCPK4 target sites in this research |
| OsCPK4-R1 | CCGAAATGGAACTCCCGAAT | |
| OsCPK7-F1 | CCTGGGAAGGATACAATTGT | Amplify the OsCPK7 target site in this research |
| OsCPK7-R1 | TAGCTATCCCCCTGAACGTA | |
| OsGSK3-F1 | TGTTCTGCCTTGCCTCACTGG | Amplify the OsGSK3 target site in this research |
| OsGSK3-R1 | ACACTGCAGAAGTTTACCTG | |
| OsGSK4-F1 | GGACAAAGAATGTCGCATGA | Amplify the OsGSK4 target site in this research |
| OsGSK4-R1 | ACATGCACAAGACTAATACG | |
| gOsMPK8-F1 | tgttgCGGAGGAAGGTGAGAGGAGG | Target the OsMPK8(20-nt)target site |
| gOsMPK8-R1 | aaacCCTCCTCTCACCTTCCTCCGc | |
| gOsMPK8-F2 | tgttgCAGACGGAGCAGCAGCAGCAGCAG | Target the OsMPK8(24-nt)target site |
| gOsMPK8-R2 | aaacCTGCTGCTGCTGCTGCTCCGTCTGc | |
| gOsWRKY45-F1 | tgttgTGGAGCTACGACGCCGTCGC | Target the OsWRKY45(20-nt)target site |
| gOsWRKY45-R1 | aaacGCGACGGCGTCGTAGCTCCAc | |
| gOsWRKY45-F2 | tgttgGCACTGGAGCTACGACGCCGTCGC | Target the OsWRKY45(24-nt)target site |
| gOsWRKY45-R2 | aaacGCGACGGCGTCGTAGCTCCAGTGCc | |
| gOsCPK6-F1 | gtgtgATGGGCAACTACTACTCGTG | Target the OsCPK6 target site |
| gOsCPK6-R1 | aaacCACGAGTAGTAGTTGCCCATc | |
| gOsCPK4-F1 | tgttGCGGGAGCGGGAAGCGGCAG | Target the OsCPK4 T1 target site |
| gOsCPK4-R1 | aaacCTGCCGCTTCCCGCTCCCGC | |
| gOsCPK4-F2 | gtgtgTCGCCGTCAAGCGCATCGAC | Target the OsCPK4 T2 target site |
| gOsCPK4-R2 | aaacGTCGATGCGCTTGACGGCGAc | |
| gOsCPK7-F1 | tgttgTGCCAGAACGGGACTCTTGG | Target the OsCPK7 target site |
| gOsCPK7-R1 | aaacCCAAGAGTCCCGTTCTGGCAc | |
| gOsGSK3-F1 | gtgtGGAAGAATGGAGAACCTAAA | Target the OsGSK3 target site |
| gOsGSK3-R1 | aaacTTTAGGTTCTCCATTCTTCC | |
| gOsGSK4-F1 | tgttgAACCTGGAATACAACACCAA | Target the OsGSK4 target site |
| gOsGSK4-R1 | aaacTTGGTGTTGTATTCCAGGTTc | |
| g4-F1 | TAAGCTTGATATCGAATTCG | Amplify the PENTR4:gRNA4 vector to construct the PENTR4:gRNA34 plasmid |
| g4-R1 | TAGCCAACACAAGCGGCAGC | |
| g34-F1 | CGCTGCCGCTTGTGTTGGCTAGGATCCATCGCAGTCAGCG | Amplify the sgRNA fragments to construct the PENTR4:gRNA34 plasmid |
| g34-R1 | CGAATTCGATATCAAGCTTATCGATACCGTCGACCTCGAG |
表1 本研究使用的引物序列
Table 1 Primer sequences used in this study
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| OsMPK8-F1 | TGGCGGTGGAGGAGAGGTGA | Amplify the OsMPK8 target site in this research |
| OsMPK8-R1 | CAACCACCCATGACCATGAG | |
| OsWRKY45-F1 | TGCAGCCGCTCAAGGTGGAG | Amplify the OsWRKY45 target site in this research |
| OsWRKY45-R1 | TCAAACCCATAATGTCGTCC | |
| OsCPK6-F1 | AGAAACAGCATCTCTTGAGG | Amplify the OsCPK6 target site in this research |
| OsCPK6-R1 | GCTACTTGTTGCTGTGCTTG | |
| OsCPK4-F1 | TTCTCATCCCACACTGCGAC | Amplify two OsCPK4 target sites in this research |
| OsCPK4-R1 | CCGAAATGGAACTCCCGAAT | |
| OsCPK7-F1 | CCTGGGAAGGATACAATTGT | Amplify the OsCPK7 target site in this research |
| OsCPK7-R1 | TAGCTATCCCCCTGAACGTA | |
| OsGSK3-F1 | TGTTCTGCCTTGCCTCACTGG | Amplify the OsGSK3 target site in this research |
| OsGSK3-R1 | ACACTGCAGAAGTTTACCTG | |
| OsGSK4-F1 | GGACAAAGAATGTCGCATGA | Amplify the OsGSK4 target site in this research |
| OsGSK4-R1 | ACATGCACAAGACTAATACG | |
| gOsMPK8-F1 | tgttgCGGAGGAAGGTGAGAGGAGG | Target the OsMPK8(20-nt)target site |
| gOsMPK8-R1 | aaacCCTCCTCTCACCTTCCTCCGc | |
| gOsMPK8-F2 | tgttgCAGACGGAGCAGCAGCAGCAGCAG | Target the OsMPK8(24-nt)target site |
| gOsMPK8-R2 | aaacCTGCTGCTGCTGCTGCTCCGTCTGc | |
| gOsWRKY45-F1 | tgttgTGGAGCTACGACGCCGTCGC | Target the OsWRKY45(20-nt)target site |
| gOsWRKY45-R1 | aaacGCGACGGCGTCGTAGCTCCAc | |
| gOsWRKY45-F2 | tgttgGCACTGGAGCTACGACGCCGTCGC | Target the OsWRKY45(24-nt)target site |
| gOsWRKY45-R2 | aaacGCGACGGCGTCGTAGCTCCAGTGCc | |
| gOsCPK6-F1 | gtgtgATGGGCAACTACTACTCGTG | Target the OsCPK6 target site |
| gOsCPK6-R1 | aaacCACGAGTAGTAGTTGCCCATc | |
| gOsCPK4-F1 | tgttGCGGGAGCGGGAAGCGGCAG | Target the OsCPK4 T1 target site |
| gOsCPK4-R1 | aaacCTGCCGCTTCCCGCTCCCGC | |
| gOsCPK4-F2 | gtgtgTCGCCGTCAAGCGCATCGAC | Target the OsCPK4 T2 target site |
| gOsCPK4-R2 | aaacGTCGATGCGCTTGACGGCGAc | |
| gOsCPK7-F1 | tgttgTGCCAGAACGGGACTCTTGG | Target the OsCPK7 target site |
| gOsCPK7-R1 | aaacCCAAGAGTCCCGTTCTGGCAc | |
| gOsGSK3-F1 | gtgtGGAAGAATGGAGAACCTAAA | Target the OsGSK3 target site |
| gOsGSK3-R1 | aaacTTTAGGTTCTCCATTCTTCC | |
| gOsGSK4-F1 | tgttgAACCTGGAATACAACACCAA | Target the OsGSK4 target site |
| gOsGSK4-R1 | aaacTTGGTGTTGTATTCCAGGTTc | |
| g4-F1 | TAAGCTTGATATCGAATTCG | Amplify the PENTR4:gRNA4 vector to construct the PENTR4:gRNA34 plasmid |
| g4-R1 | TAGCCAACACAAGCGGCAGC | |
| g34-F1 | CGCTGCCGCTTGTGTTGGCTAGGATCCATCGCAGTCAGCG | Amplify the sgRNA fragments to construct the PENTR4:gRNA34 plasmid |
| g34-R1 | CGAATTCGATATCAAGCTTATCGATACCGTCGACCTCGAG |
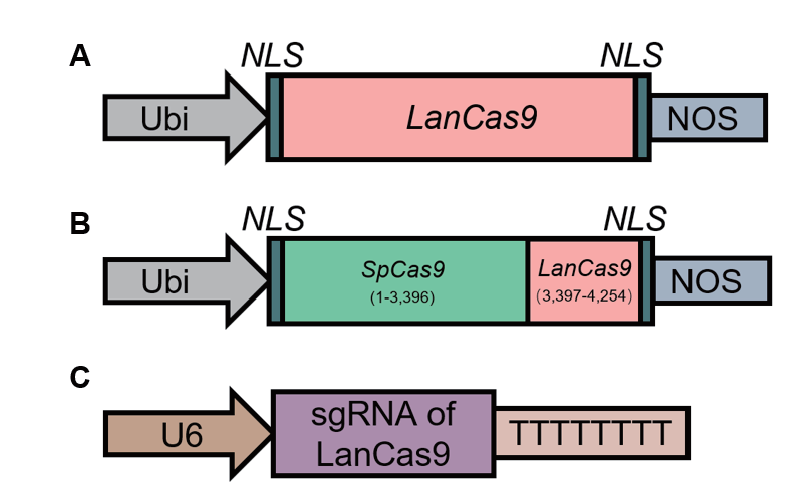
图1 载体示意图 A:LanCas9蛋白表达元件示意图;B:SLanCas9蛋白表达元件示意图;C:pENTR4-gRNA34载体示意图
Fig. 1 Schematics of vector A: Schematics of LanCas9 expression components. B: Schematics of SLanCas9 expression components. C: Schematics of pENTR4-gRNA34 vector
| 基因Gene | 基因登录号Gene identifier | 间隔区Spacer | PAM序列PAM sequence |
|---|---|---|---|
| OsWRKY45 | LOC_Os05g25770 | TGGAGCTACGACGCCGTCGC | CGG |
| TGCACTGGAGCTACGACGCCGTCGC | CGG | ||
| OsCPK4 T1 | LOC_Os02g03410 | GCGGGAGCGGGAAGCGGCAG | CAG |
| OsCPK4 T2 | LOC_Os02g03410 | TCGCCGTCAAGCGCATCGAC | AAG |
| OsCPK6 | LOC_Os02g58520 | ATGGGCAACTACTACTCGTG | CGG |
| OsCPK7 | LOC_Os03g03660 | TGCCAGAACGGGACTCTTGG | GAG |
| OsMPK8 | LOC_Os03g59390 | CGGAGGAAGGTGAGAGGAGG | TGG |
| CAGACGGAGCAGCAGCAGCAGCAG | CGG | ||
| OsGSK3 | LOC_Os02g14130 | GGAAGAATGGAGAACCTAAA | AGG |
| OsGSK4 | LOC_Os06g35530 | AACCTGGAATACAACACCAA | AGG |
表2 实验中用到的所有靶点
Table 2 Target genes in this study
| 基因Gene | 基因登录号Gene identifier | 间隔区Spacer | PAM序列PAM sequence |
|---|---|---|---|
| OsWRKY45 | LOC_Os05g25770 | TGGAGCTACGACGCCGTCGC | CGG |
| TGCACTGGAGCTACGACGCCGTCGC | CGG | ||
| OsCPK4 T1 | LOC_Os02g03410 | GCGGGAGCGGGAAGCGGCAG | CAG |
| OsCPK4 T2 | LOC_Os02g03410 | TCGCCGTCAAGCGCATCGAC | AAG |
| OsCPK6 | LOC_Os02g58520 | ATGGGCAACTACTACTCGTG | CGG |
| OsCPK7 | LOC_Os03g03660 | TGCCAGAACGGGACTCTTGG | GAG |
| OsMPK8 | LOC_Os03g59390 | CGGAGGAAGGTGAGAGGAGG | TGG |
| CAGACGGAGCAGCAGCAGCAGCAG | CGG | ||
| OsGSK3 | LOC_Os02g14130 | GGAAGAATGGAGAACCTAAA | AGG |
| OsGSK4 | LOC_Os06g35530 | AACCTGGAATACAACACCAA | AGG |
| PAM序列 PAM Seq | 基因 Gene | 打靶位点 Target site | Spacer长度 Spacer lengths/bp | 突变效率 Mutation efficiency | 单等位突变 Mono-allelic mutation | 双等位突变 Bi-allelic mutation |
|---|---|---|---|---|---|---|
| NGG | OsMPK8 | CGGAGGAAGGTGAGAGGAGGTGG | 20 | 5/48(10.42%) | 5 | 0 |
| CAGACGGAGCAGCAGCAGCAGCAGCGG | 24 | 1/48(2.08%) | 1 | 0 | ||
| OsWRKY45 | TGGAGCTACGACGCCGTCGCCGG | 20 | 12/48(25.00%) | 11 | 1 | |
| TGCACTGGAGCTACGACGCCGTCGCCGG | 24 | 2/48(4.17%) | 2 | 0 |
表3 LanCas9对水稻基因的编辑情况
Table 3 Efficiency of gene editing by LanCas9 in rice
| PAM序列 PAM Seq | 基因 Gene | 打靶位点 Target site | Spacer长度 Spacer lengths/bp | 突变效率 Mutation efficiency | 单等位突变 Mono-allelic mutation | 双等位突变 Bi-allelic mutation |
|---|---|---|---|---|---|---|
| NGG | OsMPK8 | CGGAGGAAGGTGAGAGGAGGTGG | 20 | 5/48(10.42%) | 5 | 0 |
| CAGACGGAGCAGCAGCAGCAGCAGCGG | 24 | 1/48(2.08%) | 1 | 0 | ||
| OsWRKY45 | TGGAGCTACGACGCCGTCGCCGG | 20 | 12/48(25.00%) | 11 | 1 | |
| TGCACTGGAGCTACGACGCCGTCGCCGG | 24 | 2/48(4.17%) | 2 | 0 |

图2 LanCas9诱导OsMPK8和OsWRKY45位点的编辑模式 A:用含有20 nt spacer的LanCas9打靶OsMPK8的编辑类型;B:用含有20 nt spacer的LanCas9打靶OsWRKY45的编辑类型;C:用含有24 nt spacer的LanCas9打靶OsMPK8的编辑类型;D:用含有24 nt spacer的LanCas9打靶OsWRKY45的编辑类型;PAM序列、靶序列和检测到的核苷酸改变分别以绿色、粗体和红色突出显示;WT:野生型
Fig. 2 Editing patterns of OsMPK8 and OsWRKY45 induced by LanCas9 A: Editing pattern of OsMPK8 induced by LanCas9 with 20-nt spacer. B: Editing pattern of OsWRKY45 induced by LanCas9 with 20-nt spacer. C: Editing pattern of OsMPK8 induced by LanCas9 with 24-nt spacer. D: Editing pattern of OsWRKY45 induced by LanCas9 with 24-nt spacer. The PAM sequences, target sequences, and detected nucleotide changes are highlighted in green, bold, and red, respectively; WT: wild type
| PAM序列PAM Seq | 基因Gene | 突变效率Mutation efficiency | 单等位基因突变Mono-allelic mutation | 双等位基因突变Bi-allelic mutation |
|---|---|---|---|---|
| NGG | OsWRKY45 | 25/31(80.65%) | 2 | 23 |
| OsCPK6 | 48/48(100.00%) | 5 | 43 | |
| NAG | OsCPK4 T1 | 19/48(39.58%) | 18 | 1 |
| OsCPK7 | 0/48(0.00%) | 0 | 0 |
表4 SLanCas9对水稻基因的编辑效率
Table 4 Efficiency of gene editing by SLanCas9 in rice
| PAM序列PAM Seq | 基因Gene | 突变效率Mutation efficiency | 单等位基因突变Mono-allelic mutation | 双等位基因突变Bi-allelic mutation |
|---|---|---|---|---|
| NGG | OsWRKY45 | 25/31(80.65%) | 2 | 23 |
| OsCPK6 | 48/48(100.00%) | 5 | 43 | |
| NAG | OsCPK4 T1 | 19/48(39.58%) | 18 | 1 |
| OsCPK7 | 0/48(0.00%) | 0 | 0 |
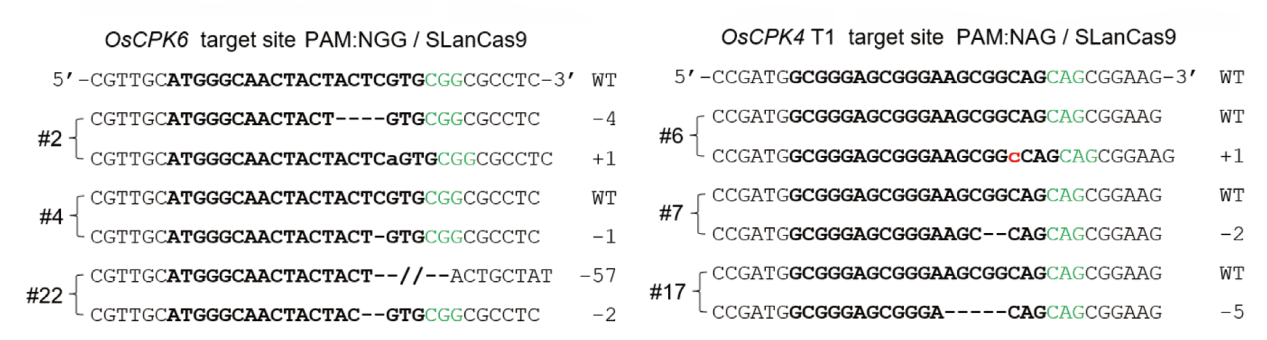
图3 SLanCas9在水稻OsCPK6和OsCPK4 T1基因中具有代表性的突变 PAM序列、靶序列和检测到的核苷酸改变分别以绿色、粗体和红色突出显示;WT:野生型
Fig. 3 Representative mutations in OsCPK6 and OsCPK4 T1 genes in rice by SLanCas9 The PAM sequences, target sequences, and detected nucleotide changes are highlighted in green, bold, and red, respectively; WT: wild type

图4 SLanCas9在水稻基因中的编辑类型及占比 Bi:双等位基因突变;Mono:单等位基因突变;Ho:纯合子;He:杂合子;Del:缺失;WT:野生型
Fig. 4 Editing type and proportion of SLanCas9 in rice's genes Bi: Bi-allelic mutation; Mono: mono-allelic mutation; Ho: homozygotes; He: heterozygotes; Del: deletion; WT: wild type
| PAM序列PAM Seq | 基因Gene | 突变效率Mutation efficiency | 单等位基因突变Mono-allelic mutation | 双等位基因突变Bi-allelic mutation |
|---|---|---|---|---|
| NGG | OsGSK3 | 21/27(77.78%) | 20 | 1 |
| OsGSK4 | 26/27(96.30%) | 10 | 16 | |
| NAG | OsCPK4 T1 | 14/48(29.17%) | 14 | 0 |
| OsCPK4 T2 | 0/48(0.00%) | 0 | 0 |
表5 SLanCas9在水稻多重基因编辑的效率
Table 5 Efficiency of SLanCas9 in multiple gene editing in rice
| PAM序列PAM Seq | 基因Gene | 突变效率Mutation efficiency | 单等位基因突变Mono-allelic mutation | 双等位基因突变Bi-allelic mutation |
|---|---|---|---|---|
| NGG | OsGSK3 | 21/27(77.78%) | 20 | 1 |
| OsGSK4 | 26/27(96.30%) | 10 | 16 | |
| NAG | OsCPK4 T1 | 14/48(29.17%) | 14 | 0 |
| OsCPK4 T2 | 0/48(0.00%) | 0 | 0 |
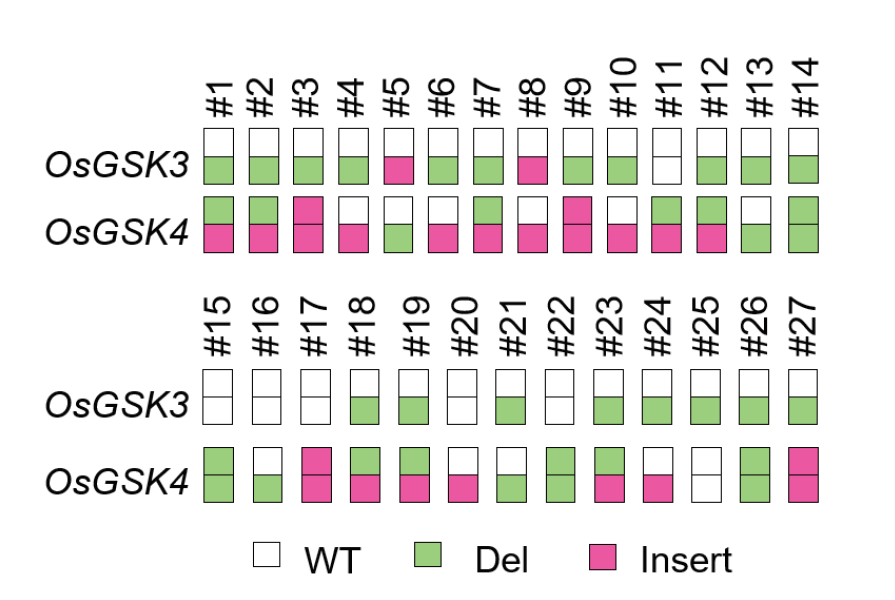
图5 SLanCas9在OsGSK3/OsGSK4多重基因编辑中的突变事件 Del:删除;Insert:插入;WT:野生型
Fig. 5 Mutational events of SLanCas9 in OsGSK3/OsGSK4 multiplex gene editing Del: Delate; Insert: insert; WT: wild type

图6 SLanCas9在OsGSK3/OsGSK4多重基因编辑中的编辑类型及占比 Bi:双等位基因突变;Mono:单等位基因突变;Ho:纯合子;He:杂合子;Del:缺失;WT:野生型
Fig. 6 The editing type and proportion of SLanCas9 in OsGSK3/OsGSK4 multiple gene editing Bi: Bi-allelic mutation; Mono: mono-allelic mutation; Ho: homozygotes; He: heterozygotes; Del: deletion; WT: wild type

图7 SLanCas9在OsCPK4 T1多重基因编辑中具有代表性的突变 PAM序列、靶序列和检测到的核苷酸改变分别以绿色、粗体和红色突出显示;WT:野生型
Fig. 7 Representative mutation in OsCPK4 T1 multiplex gene editing of SLanCas9 The PAM sequences, target sequences, and detected nucleotide changes are highlighted in green, bold, and red, respectively; WT: wild type
| [1] |
Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821.
doi: 10.1126/science.1225829 pmid: 22745249 |
| [2] |
Mali P, Yang LH, Esvelt KM, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826.
doi: 10.1126/science.1232033 pmid: 23287722 |
| [3] |
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nat Biotechnol, 2020, 38(7): 824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [4] |
Yan DQ, Ren B, Liu L, et al. High-efficiency and multiplex adenine base editing in plants using new TadA variants[J]. Mol Plant, 2021, 14(5): 722-731.
doi: 10.1016/j.molp.2021.02.007 pmid: 33631420 |
| [5] |
Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases[J]. Nat Rev Genet, 2010, 11(9): 636-646.
doi: 10.1038/nrg2842 pmid: 20717154 |
| [6] | Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing[J]. Nat Rev Mol Cell Biol, 2013, 14(1): 49-55. |
| [7] |
Shan QW, Wang YP, Li J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system[J]. Nat Biotechnol, 2013, 31(8): 686-688.
doi: 10.1038/nbt.2650 pmid: 23929338 |
| [8] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278.
doi: S0092-8674(14)00604-7 pmid: 24906146 |
| [9] | 尤李兰, 孙伟, 杨晓琪, 等. 诺贝尔化学奖授予CRISPR-Cas9基因编辑研究[J]. 生物化学与生物物理进展, 2020, 47(11): 1119-1126. |
| You LL, Sun W, Yang XQ, et al. Chemistry Nobel honors CRISPR-Cas9[J]. Prog Biochem Biophys, 2020, 47(11): 1119-1126. | |
| [10] | Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR-CAS systems[J]. Biochem Soc Trans, 2013, 41(6): 1392-1400. |
| [11] | Haft DH, Selengut J, Mongodin EF, et al. A guild of 45 CRISPR-associated(Cas)protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes[J]. PLoS Comput Biol, 2005, 1(6): e60. |
| [12] |
Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. J Bacteriol, 1987, 169(12): 5429-5433.
doi: 10.1128/jb.169.12.5429-5433.1987 pmid: 3316184 |
| [13] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [14] | Gutási A, Hammer SE, El-Matbouli M, et al. Review: recent applications of gene editing in fish species and aquatic medicine[J]. Animals, 2023, 13(7): 1250. |
| [15] |
Ren B, Liu L, Li SF, et al. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice[J]. Mol Plant, 2019, 12(7): 1015-1026.
doi: S1674-2052(19)30123-6 pmid: 30928635 |
| [16] | Yan F, Wang JW, Zhang SJ, et al. CRISPR/FnCas12a-mediated efficient multiplex and iterative genome editing in bacterial plant pathogens without donor DNA templates[J]. PLoS Pathog, 2023, 19(1): e1010961. |
| [17] |
Ceasar SA, Rajan V, Prykhozhij SV, et al. Insert, remove or replace: a highly advanced genome editing system using CRISPR/Cas9[J]. Biochim Biophys Acta, 2016, 1863(9): 2333-2344.
doi: 10.1016/j.bbamcr.2016.06.009 pmid: 27350235 |
| [18] |
Hille F, Richter H, Wong SP, et al. The biology of CRISPR-cas: backward and forward[J]. Cell, 2018, 172(6): 1239-1259.
doi: S0092-8674(17)31383-1 pmid: 29522745 |
| [19] |
Gasiunas G, Young JK, Karvelis T, et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs[J]. Nat Commun, 2020, 11: 5512.
doi: 10.1038/s41467-020-19344-1 pmid: 33139742 |
| [20] | Wang MX, Yan F, Zhou HB. Protocol for targeted modification of the rice genome using base editing[J]. STAR Protoc, 2022, 3(4): 101865. |
| [21] | Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J]. Plant Mol Biol Report, 1997, 15(1): 8-15. |
| [22] |
Hua K, Tao XP, Han PJ, et al. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences[J]. Mol Plant, 2019, 12(7): 1003-1014.
doi: S1674-2052(19)30122-4 pmid: 30928636 |
| [23] |
Li JY, Luo JM, Xu ML, et al. Plant genome editing using xCas9 with expanded PAM compatibility[J]. J Genet Genomics, 2019, 46(5): 277-280.
doi: S1673-8527(19)30052-9 pmid: 31054950 |
| [24] |
Wang JJ, Meng XB, Hu XX, et al. xCas9 expands the scope of genome editing with reduced efficiency in rice[J]. Plant Biotechnol J, 2019, 17(4): 709-711.
doi: 10.1111/pbi.13053 pmid: 30549238 |
| [25] |
Xu YB, Meng XB, Wang JJ, et al. ScCas9 recognizes NNG protospacer adjacent motif in genome editing of rice[J]. Sci China Life Sci, 2020, 63(3): 450-452.
doi: 10.1007/s11427-019-1630-2 pmid: 31953707 |
| [26] |
Ren J, Meng XB, Hu FY, et al. Expanding the scope of genome editing with SpG and SpRY variants in rice[J]. Sci China Life Sci, 2021, 64(10): 1784-1787.
doi: 10.1007/s11427-020-1883-5 pmid: 33443621 |
| [27] | Huang JY, Lin QP, Fei HY, et al. Discovery of deaminase functions by structure-based protein clustering[J]. Cell, 2023, 186(15): 3182-3195.e14. |
| [28] |
Tsuchida CA, Zhang SY, Doost MS, et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity[J]. Mol Cell, 2022, 82(6): 1199-1209.e6.
doi: 10.1016/j.molcel.2022.02.002 pmid: 35219382 |
| [29] |
Xu ZY, Kuang YJ, Ren B, et al. SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition[J]. Genome Biol, 2021, 22(1): 6.
doi: 10.1186/s13059-020-02231-9 pmid: 33397431 |
| [30] |
Chen KL, Wang YP, Zhang R, et al. CRISPR/cas genome editing and precision plant breeding in agriculture[J]. Annu Rev Plant Biol, 2019, 70: 667-697.
doi: 10.1146/annurev-arplant-050718-100049 pmid: 30835493 |
| [1] | 刘文志, 贺丹, 李鹏, 傅应林, 张译心, 温华杰, 于文清. 多粘类芽胞杆菌新菌株X-11及其对番茄和水稻的促生效应[J]. 生物技术通报, 2024, 40(9): 249-259. |
| [2] | 童玮婧, 罗数, 陆新露, 沈建福, 陆柏益, 李开绵, 马秋香, 张鹏. CRISPR/Cas9编辑MeHNL基因创制低生氰糖苷木薯[J]. 生物技术通报, 2024, 40(9): 11-19. |
| [3] | 朱诗斐, 刘敬, 张家芊, 黄文坤, 彭德良, 孔令安, 彭焕. 水稻和拟禾本科根结线虫互作分子机制研究进展[J]. 生物技术通报, 2024, 40(9): 172-180. |
| [4] | 崔海洋, 谭淼, 全壮, 陈红利, 董艳敏, 唐立春. 利用Cas9TX实现非病毒TRAC定点整合制备T细胞[J]. 生物技术通报, 2024, 40(9): 190-197. |
| [5] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [6] | 孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62. |
| [7] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [8] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [9] | 隆静, 陈婧敏, 刘霄, 张一凡, 周利斌, 杜艳. 植物DNA双链断裂修复机制及其在重离子诱变和基因编辑中的作用[J]. 生物技术通报, 2024, 40(7): 55-67. |
| [10] | 陈墨岩, 祝诚. 基于CRISPR/Cas12a的生物传感平台的机制研究及应用[J]. 生物技术通报, 2024, 40(7): 90-98. |
| [11] | 田胜尼, 张琴, 董玉飞, 丁洲, 叶爱华, 张明珠. 酸性矿山废水对成熟期水稻根区理化因子及固氮微生物的影响[J]. 生物技术通报, 2024, 40(6): 271-280. |
| [12] | 肖怡梦, 杨雯, 程依依, 罗刚. CRISPR-Cas9基因编辑技术及其在家禽中的研究进展[J]. 生物技术通报, 2024, 40(5): 38-47. |
| [13] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [14] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [15] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||