








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 41-52.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0638
收稿日期:2024-07-05
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
武志强,男,博士,研究员,研究方向:植物细胞器基因组;E-mail: wuzhiqiang@caas.cn作者简介:张硕,男,博士研究生,研究方向:植物线粒体基因组编辑;E-mail: szhang@webmail.hzau.edu.cn
基金资助:
ZHANG Shuo1,2( ), KAN Jun-hu1, ZHOU Jia-wei1, WU Zhi-qiang1(
), KAN Jun-hu1, ZHOU Jia-wei1, WU Zhi-qiang1( )
)
Received:2024-07-05
Published:2024-10-26
Online:2024-11-20
摘要:
线粒体是真核生物细胞内的半自主细胞器,具有自身的基因组(mtDNA),在生命活动中扮演重要角色。人类mtDNA突变与多种遗传疾病相关,而在植物中mtDNA高频重组产生的ORF基因常常与植株雄性不育表型相关。随着基因编辑技术的迅速发展,mtDNA编辑技术为研究和治疗线粒体疾病提供了有力工具。得益于mtDNA编辑工具的丰富,线粒体编辑技术也被广泛应用于植物线粒体基因组功能基因以及未知序列的研究中。相较于核基因组编辑,针对mtDNA编辑仍然面临一些限制因素。本文总结了mtDNA编辑技术的发展和研究现状,以及在植物领域利用这些编辑技术对mtDNA进行的研究进展,展望了在植物线粒体研究中的mtDNA编辑技术潜在优化思路以及应用潜力。
张硕, 阚俊虎, 周家伟, 武志强. 植物线粒体基因组编辑研究进展[J]. 生物技术通报, 2024, 40(10): 41-52.
ZHANG Shuo, KAN Jun-hu, ZHOU Jia-wei, WU Zhi-qiang. Advance in Plant Mitochondrial Genome Editing[J]. Biotechnology Bulletin, 2024, 40(10): 41-52.
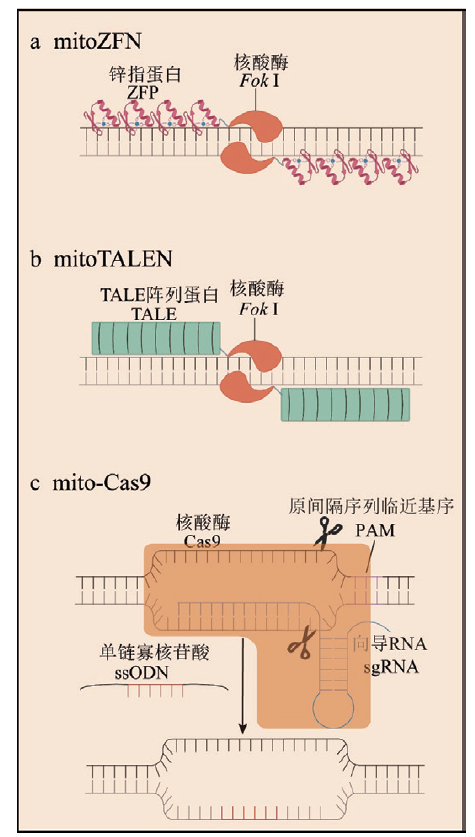
图1 线粒体基因组编辑技术示意图 a: mitoZFN中串联的ZFP锌指蛋白特异识别靶序列,Fok I形成的二聚体具有核酸酶活性;b: mitoTALEN中串联的TALE蛋白特异识别靶序列,Fok I形成的二聚体具有核酸酶活性;c:mito-Cas9中Cas9蛋白可以借助MTS进入线粒体内,而sgRNA和单链寡核苷酸只能有限的进入线粒体内,限制了mito-Cas9的编辑效率
Fig. 1 Schematic diagram of mitochondrial genome editing technology a: In mitoZFN, tandem ZFP zinc finger proteins specifically recognize target sequences, and the Fok I dimer exhibits nuclease activity. b: In mitoTALEN, tandem TALE proteins specifically recognize target sequences, and the Fok I dimer exhibits nuclease activity. c: In mito-Cas9, the Cas9 protein can enter the mitochondria with the help of MTS, while sgRNA and single-stranded oligonucleotides can only enter the mitochondria to a limited extent, restricting the editing efficiency of mito-Cas9

图2 在植物中应用的线粒体基因组编辑技术示意图 a: mitoTALEN经过密码子优后应用到植物线粒体研究中;b: mt-DdCBEs/mitoTALECD利用串联的TALE蛋白特异识别靶序列,DddA脱氨酶将胞嘧啶转化为尿嘧啶,UGI作为尿嘧啶糖基化酶抑制剂保护尿嘧啶,经过DNA复制实现C-to-T的编辑;c:TALED中利用DddA脱氨酶打开双链DNA,具有单链特异性TadA8e腺嘌呤脱氨酶将腺嘌呤转化为次黄嘌呤,经过DNA复制实现A-to-G的编辑;d:mitoCRISPR/Cas9中Cas9蛋白借助MTS进入线粒体内与sgRNA结合实现对靶序列的切割
Fig. 2 Schematic representation of mitochondrial genome editing techniques applied in plants a: Codon-optimized mitoTALEN has been applied to plant mitochondrial research. b: mt-DdCBEs/mitoTALECD utilizes tandem TALE proteins to specifically recognize target sequences. The DddA deaminase converts cytosine to uracil, and UGI acts as a uracil glycosylase inhibitor to protect uracil, achieving C-to-T editing through DNA replication. c: In TALED, DddA deaminase opens double-stranded DNA, and the single-strand-specific TadA8e adenine deaminase converts adenine to inosine, achieving A-to-G editing through DNA replication. d: In mitoCRISPR/Cas9, the Cas9 protein, aided by MTS, enters the mitochondria and binds with sgRNA to achieve target sequence cleavage
| 编辑工具 Editing tool | 物种 Species | 靶基因 Target gene | 编辑类型 Editing type | 表型 Phenotype | 文献 Reference |
|---|---|---|---|---|---|
| mitoTALEN | 水稻Oryza sativa | orf79 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 油菜 Brassica napus | orf125 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 拟南芥Arabidopsis thaliana | atp6-1, atp6-2 | 敲除 | 功能冗余;双突致死 | [ |
| mitoTALEN | 水稻Oryza sativa | orf352 | 敲除 | 恢复花粉育性;不结实 | [ |
| mitoTALEN | 水稻Oryza sativa | orf312 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 番茄Solanum lycopersicum | orf137 | 敲除 | 恢复花粉育性 | [ |
| TALEN-GDM | 烟草 Nicotiana tabacum | nad9 | 点突变 | 无明显表型 | [ |
| TALEN-GDM | 烟草Nicotiana tabacum | nad9 | 敲除 | 生长迟缓;叶片和花发育缺陷;雄性不育 | [ |
| mitoTALEN | 拟南芥Arabidopsis thaliana | nad7 | 敲除 | 严重的生长迟缓;致死 | [ |
| mitoTALEN | 水稻Oryza sativa | WA352 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 西兰花Brassica oleracea | orf138 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 马铃薯Solanum tuberosum | orf125 | 敲除 | N/A | [ |
| mt-DdCBEs | 油菜Brassica napus | atp6, rps14 | C-to-T | N/A | [ |
| mt-DdCBEs | 莴苣Lactuca sativa | atp6 | C-to-T | N/A | [ |
| mitoTALECD | 拟南芥Arabidopsis thaliana | atp1 | C-to-T | 恢复正常生长 | [ |
| mitoTALECD | 马铃薯Solanum tuberosum | orf125 | C-to-T | N/A | [ |
| mTALED | 水稻Oryza sativa | atp6-2 | A-to-G | N/A | [ |
| mitoCRISPR/Cas9 | 烟草Nicotiana tabacum | atp9 | 敲除 | 雄性不育 | [ |
表1 线粒体基因组编辑技术在植物线粒体基因功能研究中的应用
Table 1 Application of mitochondrial genome editing technology in the study of plant mitochondrial gene functions
| 编辑工具 Editing tool | 物种 Species | 靶基因 Target gene | 编辑类型 Editing type | 表型 Phenotype | 文献 Reference |
|---|---|---|---|---|---|
| mitoTALEN | 水稻Oryza sativa | orf79 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 油菜 Brassica napus | orf125 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 拟南芥Arabidopsis thaliana | atp6-1, atp6-2 | 敲除 | 功能冗余;双突致死 | [ |
| mitoTALEN | 水稻Oryza sativa | orf352 | 敲除 | 恢复花粉育性;不结实 | [ |
| mitoTALEN | 水稻Oryza sativa | orf312 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 番茄Solanum lycopersicum | orf137 | 敲除 | 恢复花粉育性 | [ |
| TALEN-GDM | 烟草 Nicotiana tabacum | nad9 | 点突变 | 无明显表型 | [ |
| TALEN-GDM | 烟草Nicotiana tabacum | nad9 | 敲除 | 生长迟缓;叶片和花发育缺陷;雄性不育 | [ |
| mitoTALEN | 拟南芥Arabidopsis thaliana | nad7 | 敲除 | 严重的生长迟缓;致死 | [ |
| mitoTALEN | 水稻Oryza sativa | WA352 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 西兰花Brassica oleracea | orf138 | 敲除 | 恢复花粉育性 | [ |
| mitoTALEN | 马铃薯Solanum tuberosum | orf125 | 敲除 | N/A | [ |
| mt-DdCBEs | 油菜Brassica napus | atp6, rps14 | C-to-T | N/A | [ |
| mt-DdCBEs | 莴苣Lactuca sativa | atp6 | C-to-T | N/A | [ |
| mitoTALECD | 拟南芥Arabidopsis thaliana | atp1 | C-to-T | 恢复正常生长 | [ |
| mitoTALECD | 马铃薯Solanum tuberosum | orf125 | C-to-T | N/A | [ |
| mTALED | 水稻Oryza sativa | atp6-2 | A-to-G | N/A | [ |
| mitoCRISPR/Cas9 | 烟草Nicotiana tabacum | atp9 | 敲除 | 雄性不育 | [ |
| [1] |
Sloan DB, Warren JM, Williams AM, et al. Cytonuclear integration and co-evolution[J]. Nat Rev Genet, 2018, 19(10): 635-648.
doi: 10.1038/s41576-018-0035-9 pmid: 30018367 |
| [2] | Wang J, Kan SL, Liao XZ, et al. Plant organellar genomes: much done, much more to do[J]. Trends Plant Sci, 2024, 29(7): 754-769. |
| [3] |
Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture[J]. Science, 2006, 311(5768): 1727-1730.
doi: 10.1126/science.1118884 pmid: 16556832 |
| [4] | Hu ZJ, Yang L, Zhang ML, et al. A novel protein CYTB-187AA encoded by the mitochondrial gene CYTB modulates mammalian early development[J]. Cell Metab, 2024, 36(7): 1586-1597.e7. |
| [5] |
Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy[J]. Science, 1988, 242(4884): 1427-1430.
doi: 10.1126/science.3201231 pmid: 3201231 |
| [6] |
Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: an update[J]. Cell Metab, 2017, 25(1): 57-71.
doi: S1550-4131(16)30502-2 pmid: 28094012 |
| [7] | Yang L, Lin XB, Tang HT, et al. Mitochondrial DNA mutation exacerbates female reproductive aging via impairment of the NADH/NAD+ redox[J]. Aging Cell, 2020, 19(9): e13206. |
| [8] |
DiMauro S, Hirano M, Schon EA. Approaches to the treatment of mitochondrial diseases[J]. Muscle Nerve, 2006, 34(3): 265-283.
pmid: 16810684 |
| [9] |
Zhang J, Liu H, Luo SY, et al. Live birth derived from oocyte spindle transfer to prevent mitochondrial disease[J]. Reprod Biomed Online, 2017, 34(4): 361-368.
doi: S1472-6483(17)30041-X pmid: 28385334 |
| [10] |
Minczuk M, Papworth MA, Miller JC, et al. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA[J]. Nucleic Acids Res, 2008, 36(12): 3926-3938.
doi: 10.1093/nar/gkn313 pmid: 18511461 |
| [11] | Kim JS, Chen J. Base editing of organellar DNA with programmable deaminases[J]. Nat Rev Mol Cell Biol, 2024, 25(1): 34-45. |
| [12] | Kozik A, Rowan BA, Lavelle D, et al. The alternative reality of plant mitochondrial DNA: one ring does not rule them all[J]. PLoS Genet, 2019, 15(8): e1008373. |
| [13] | Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence[J]. J Mol Evol, 1988, 28(1/2): 87-97. |
| [14] |
Christensen AC. Plant mitochondria are a riddle wrapped in a mystery inside an Enigma[J]. J Mol Evol, 2021, 89(3): 151-156.
doi: 10.1007/s00239-020-09980-y pmid: 33486550 |
| [15] |
Chen LT, Liu YG. Male sterility and fertility restoration in crops[J]. Annu Rev Plant Biol, 2014, 65: 579-606.
doi: 10.1146/annurev-arplant-050213-040119 pmid: 24313845 |
| [16] |
Luo DP, Xu H, Liu ZL, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice[J]. Nat Genet, 2013, 45(5): 573-577.
doi: 10.1038/ng.2570 pmid: 23502780 |
| [17] |
Minczuk M, Papworth MA, Kolasinska P, et al. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase[J]. Proc Natl Acad Sci USA, 2006, 103(52): 19689-19694.
doi: 10.1073/pnas.0609502103 pmid: 17170133 |
| [18] |
Bacman SR, Williams SL, Pinto M, et al. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs[J]. Nat Med, 2013, 19(9): 1111-1113.
doi: 10.1038/nm.3261 pmid: 23913125 |
| [19] | Zhu HC, Li C, Gao CX. Applications of CRISPR-Cas in agriculture and plant biotechnology[J]. Nat Rev Mol Cell Biol, 2020, 21(11): 661-677. |
| [20] |
Pacesa M, Pelea O, Jinek M. Past, present, and future of CRISPR genome editing technologies[J]. Cell, 2024, 187(5): 1076-1100.
doi: 10.1016/j.cell.2024.01.042 pmid: 38428389 |
| [21] | Bi R, Li Y, Xu M, et al. Direct evidence of CRISPR-Cas9-mediated mitochondrial genome editing[J]. Innovation(Camb), 2022, 3(6): 100329. |
| [22] |
Gammage PA, Moraes CT, Minczuk M. Mitochondrial genome engineering: the revolution may not be CRISPR-ized[J]. Trends Genet, 2018, 34(2): 101-110.
doi: S0168-9525(17)30191-9 pmid: 29179920 |
| [23] |
Beerli RR, Barbas CF III. Engineering polydactyl zinc-finger transcription factors[J]. Nat Biotechnol, 2002, 20(2): 135-141.
pmid: 11821858 |
| [24] |
Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease[J]. Proc Natl Acad Sci USA, 1992, 89(10): 4275-4279.
pmid: 1584761 |
| [25] |
Gammage PA, Rorbach J, Vincent AI, et al. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations[J]. EMBO Mol Med, 2014, 6(4): 458-466.
doi: 10.1002/emmm.201303672 pmid: 24567072 |
| [26] |
Lim K, Cho SI, Kim JS. Nuclear and mitochondrial DNA editing in human cells with zinc finger deaminases[J]. Nat Commun, 2022, 13(1): 366.
doi: 10.1038/s41467-022-27962-0 pmid: 35042880 |
| [27] |
Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases[J]. Nat Rev Genet, 2010, 11(9): 636-646.
doi: 10.1038/nrg2842 pmid: 20717154 |
| [28] |
Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors[J]. Science, 2009, 326(5959): 1501.
doi: 10.1126/science.1178817 pmid: 19933106 |
| [29] | Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function[J]. Annu Rev Phytopathol, 2010, 48: 419-436. |
| [30] |
Sakuma T, Ochiai H, Kaneko T, et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity[J]. Sci Rep, 2013, 3: 3379.
doi: 10.1038/srep03379 pmid: 24287550 |
| [31] |
Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors[J]. Science, 2009, 326(5959): 1509-1512.
doi: 10.1126/science.1178811 pmid: 19933107 |
| [32] | Bacman SR, Barrera-Paez JD, Pinto M, et al. mitoTALEN reduces the mutant mtDNA load in neurons[J]. Mol Ther Nucleic Acids, 2024, 35(1): 102132. |
| [33] | Bacman SR, Kauppila JHK, Pereira CV, et al. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation[J]. Nat Med, 2018, 24(11): 1696-1700. |
| [34] | Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| [35] | Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| [36] | Mok BY, de Moraes MH, Zeng J, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing[J]. Nature, 2020, 583(7817): 631-637. |
| [37] |
Qi XL, Chen XX, Guo JY, et al. Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing[J]. Cell Discov, 2021, 7(1): 95.
doi: 10.1038/s41421-021-00325-7 pmid: 34663794 |
| [38] |
Wei YH, Xu CL, Feng H, et al. Human cleaving embryos enable efficient mitochondrial base-editing with DdCBE[J]. Cell Discov, 2022, 8(1): 7.
doi: 10.1038/s41421-021-00372-0 pmid: 35102133 |
| [39] |
Lee H, Lee S, Baek G, et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases[J]. Nat Commun, 2021, 12(1): 1190.
doi: 10.1038/s41467-021-21464-1 pmid: 33608520 |
| [40] | Guo JY, Chen XX, Liu ZW, et al. DdCBE mediates efficient and inheritable modifications in mouse mitochondrial genome[J]. Mol Ther Nucleic Acids, 2021, 27: 73-80. |
| [41] | Lei ZX, Meng HW, Liu LL, et al. Mitochondrial base editor induces substantial nuclear off-target mutations[J]. Nature, 2022, 606(7915): 804-811. |
| [42] |
Wei YH, Li ZF, Xu K, et al. Mitochondrial base editor DdCBE causes substantial DNA off-target editing in nuclear genome of embryos[J]. Cell Discov, 2022, 8(1): 27.
doi: 10.1038/s41421-022-00391-5 pmid: 35304438 |
| [43] |
Mok BY, Kotrys AV, Raguram A, et al. CRISPR-free base editors with enhanced activity and expanded targeting scope in mitochondrial and nuclear DNA[J]. Nat Biotechnol, 2022, 40(9): 1378-1387.
doi: 10.1038/s41587-022-01256-8 pmid: 35379961 |
| [44] | Lee S, Lee H, Baek G, et al. Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors[J]. Nat Biotechnol, 2023, 41(3): 378-386. |
| [45] |
Sun HF, Wang ZJ, Shen L, et al. Developing mitochondrial base editors with diverse context compatibility and high fidelity via saturated spacer library[J]. Nat Commun, 2023, 14(1): 6625.
doi: 10.1038/s41467-023-42359-3 pmid: 37857619 |
| [46] | Cho SI, Lee S, Mok YG, et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases[J]. Cell, 2022, 185(10): 1764-1776.e12. |
| [47] | Cho SI, Lim K, Hong S, et al. Engineering TALE-linked deaminases to facilitate precision adenine base editing in mitochondrial DNA[J]. Cell, 2024, 187(1): 95-109.e26. |
| [48] | Hu JC, Sun Y, Li BS, et al. Strand-preferred base editing of organellar and nuclear genomes using CyDENT[J]. Nat Biotechnol, 2024, 42(6): 936-945. |
| [49] | Yi ZY, Zhang XX, Tang W, et al. Strand-selective base editing of human mitochondrial DNA using mitoBEs[J]. Nat Biotechnol, 2024, 42(3): 498-509. |
| [50] | Li SY, Xia LQ. Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives[J]. aBIOTECH, 2019, 1(1): 58-73. |
| [51] |
Gao CX. Genome engineering for crop improvement and future agriculture[J]. Cell, 2021, 184(6): 1621-1635.
doi: 10.1016/j.cell.2021.01.005 pmid: 33581057 |
| [52] | Ma GG, Kuang YJ, Lu ZW, et al. CRISPR/Sc++-mediated genome editing in rice[J]. J Integr Plant Biol, 2021, 63(9): 1606-1610. |
| [53] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278.
doi: S0092-8674(14)00604-7 pmid: 24906146 |
| [54] | Jo A, Ham S, Lee GH, et al. Efficient mitochondrial genome editing by CRISPR/Cas9[J]. Biomed Res Int, 2015, 2015: 305716. |
| [55] | Bian WP, Chen YL, Luo JJ, et al. Knock-In strategy for editing human and zebrafish mitochondrial DNA using mito-CRISPR/Cas9 system[J]. ACS Synth Biol, 2019, 8(4): 621-632. |
| [56] | Yoo BC, Yadav NS, Orozco EM Jr, et al. Cas9/gRNA-mediated genome editing of yeast mitochondria and Chlamydomonas chloroplasts[J]. PeerJ, 2020, 8: e8362. |
| [57] |
Johnston SA, Anziano PQ, Shark K, et al. Mitochondrial transformation in yeast by bombardment with microprojectiles[J]. Science, 1988, 240(4858): 1538-1541.
pmid: 2836954 |
| [58] |
Remacle C, Cardol P, Coosemans N, et al. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes[J]. Proc Natl Acad Sci USA, 2006, 103(12): 4771-4776.
pmid: 16537419 |
| [59] |
Li WM, Ruf S, Bock R. Chloramphenicol acetyltransferase as selectable marker for plastid transformation[J]. Plant Mol Biol, 2011, 76(3-5): 443-451.
doi: 10.1007/s11103-010-9678-4 pmid: 20721602 |
| [60] | Lin JY, Liu YC, Tseng YH, et al. TALE-based organellar genome editing and gene expression in plants[J]. Plant Cell Rep, 2024, 43(3): 61. |
| [61] |
Kazama T, Okuno M, Watari Y, et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing[J]. Nat Plants, 2019, 5(7): 722-730.
doi: 10.1038/s41477-019-0459-z pmid: 31285556 |
| [62] |
Omukai S, Arimura SI, Toriyama K, et al. Disruption of mitochondrial open reading frame 352 partially restores pollen development in cytoplasmic male sterile rice[J]. Plant Physiol, 2021, 187(1): 236-246.
doi: 10.1093/plphys/kiab236 pmid: 34015134 |
| [63] | Takatsuka A, Kazama T, Arimura SI, et al. TALEN-mediated depletion of the mitochondrial gene orf312 proves that it is a Tadukan-type cytoplasmic male sterility-causative gene in rice[J]. Plant J, 2022, 110(4): 994-1004. |
| [64] | Zhou JW, Nie LY, Zhang S, et al. Mitochondrial genome editing of WA352 via mitoTALENs restore fertility in cytoplasmic male sterile rice[J]. Plant Biotechnol J, 2024, 22(7): 1960-1962. |
| [65] |
Kuwabara K, Arimura SI, Shirasawa K, et al. orf137 triggers cytoplasmic male sterility in tomato[J]. Plant Physiol, 2022, 189(2): 465-468.
doi: 10.1093/plphys/kiac082 pmid: 35212743 |
| [66] |
Xu FY, Su TB, Zhang XC, et al. Editing of ORF138 restores fertility of Ogura cytoplasmic male sterile broccoli via mitoTALENs[J]. Plant Biotechnol J, 2024, 22(5): 1325-1334.
doi: 10.1111/pbi.14268 pmid: 38213067 |
| [67] | Arimura SI, Nakazato I. Genome editing of plant mitochondrial and chloroplast genomes[J]. Plant Cell Physiol, 2024, 65(4): 477-483. |
| [68] | Møller IM, Rasmusson AG, Van Aken O. Plant mitochondria - past, present and future[J]. Plant J, 2021, 108(4): 912-959. |
| [69] | Arimura SI, Ayabe H, Sugaya H, et al. Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabi-dopsis thaliana by mitoTALENs[J]. Plant J, 2020, 104(6): 1459-1471. |
| [70] | Ayabe H, Toyoda A, Iwamoto A, et al. Mitochondrial gene defects in Arabidopsis can broadly affect mitochondrial gene expression through copy number[J]. Plant Physiol, 2023, 191(4): 2256-2275. |
| [71] |
Forner J, Kleinschmidt D, Meyer EH, et al. Targeted introduction of heritable point mutations into the plant mitochondrial genome[J]. Nat Plants, 2022, 8(3): 245-256.
doi: 10.1038/s41477-022-01108-y pmid: 35301443 |
| [72] |
Forner J, Kleinschmidt D, Meyer EH, et al. Targeted knockout of a conserved plant mitochondrial gene by genome editing[J]. Nat Plants, 2023, 9(11): 1818-1831.
doi: 10.1038/s41477-023-01538-2 pmid: 37814021 |
| [73] | Kang BC, Bae SJ, Lee S, et al. Chloroplast and mitochondrial DNA editing in plants[J]. Nat Plants, 2021, 7(7): 899-905. |
| [74] | Nakazato I, Okuno M, Zhou C, et al. Targeted base editing in the mitochondrial genome of Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2022, 119(20): e2121177119. |
| [75] |
Small ID, Schallenberg-Rüdinger M, Takenaka M, et al. Plant organellar RNA editing: what 30 years of research has revealed[J]. Plant J, 2020, 101(5): 1040-1056.
doi: 10.1111/tpj.14578 |
| [76] | Small I, Melonek J, Bohne AV, et al. Plant organellar RNA maturation[J]. Plant Cell, 2023, 35(6): 1727-1751. |
| [77] | Zhou C, Okuno M, Nakazato I, et al. Targeted A-to-G base editing in the organellar genomes of Arabidopsis with monomeric programmable deaminases[J]. Plant Physiol, 2024, 194(4): 2278-2287. |
| [78] | Chang YZ, Liu BL, Jiang YY, et al. Induce male sterility by CRISPR/Cas9-mediated mitochondrial genome editing in tobacco[J]. Funct Integr Genomics, 2023, 23(3): 205. |
| [79] |
Nicolia A, Scotti N, D'Agostino N, et al. Mitochondrial DNA editing in potato through mitoTALEN and mitoTALECD: molecular characterization and stability of editing events[J]. Plant Methods, 2024, 20(1): 4.
doi: 10.1186/s13007-023-01124-9 pmid: 38183104 |
| [80] |
Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease[J]. Hum Mol Genet, 2001, 10(26): 3093-3099.
pmid: 11751691 |
| [81] |
Tanaka M, Borgeld HJ, Zhang J, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria[J]. J Biomed Sci, 2002, 9(6 Pt 1):534-541.
doi: 10.1159/000064726 pmid: 12372991 |
| [82] |
Bayona-Bafaluy MP, Blits B, Battersby BJ, et al. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease[J]. Proc Natl Acad Sci USA, 2005, 102(40): 14392-14397.
pmid: 16179392 |
| [83] |
Lee S, Lee H, Baek G, et al. Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases[J]. Genome Biol, 2022, 23(1): 211.
doi: 10.1186/s13059-022-02782-z pmid: 36224582 |
| [84] | Xu K, Feng H, Zhang HH, et al. Structure-guided discovery of highly efficient cytidine deaminases with sequence-context independence[J]. Nat Biomed Eng, 2024. |
| [85] |
Wang G, Chen HW, Oktay Y, et al. PNPASE regulates RNA import into mitochondria[J]. Cell, 2010, 142(3): 456-467.
doi: 10.1016/j.cell.2010.06.035 pmid: 20691904 |
| [86] | Wang PC, Zhang LX, Chen SY, et al. ANT2 functions as a translocon for mitochondrial cross-membrane translocation of RNAs[J]. Cell Res, 2024, 34(7): 504-521. |
| [87] | 王熠晨, 王颖, 陈妤, 等. 线粒体基因编辑技术研究进展[J]. 浙江大学学报: 医学版, 2023, 52(4): 460-472. |
| Wang YC, Wang Y, Chen Y, et al. Research progress in mitochondrial gene editing technology[J]. J Zhejiang Univ Med Sci, 2023, 52(4): 460-472. | |
| [88] |
Kolesnikova O, Kazakova H, Comte C, et al. Selection of RNA aptamers imported into yeast and human mitochondria[J]. RNA, 2010, 16(5): 926-941.
doi: 10.1261/rna.1914110 pmid: 20348443 |
| [1] | 童玮婧, 罗数, 陆新露, 沈建福, 陆柏益, 李开绵, 马秋香, 张鹏. CRISPR/Cas9编辑MeHNL基因创制低生氰糖苷木薯[J]. 生物技术通报, 2024, 40(9): 11-19. |
| [2] | 崔海洋, 谭淼, 全壮, 陈红利, 董艳敏, 唐立春. 利用Cas9TX实现非病毒TRAC定点整合制备T细胞[J]. 生物技术通报, 2024, 40(9): 190-197. |
| [3] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [4] | 隆静, 陈婧敏, 刘霄, 张一凡, 周利斌, 杜艳. 植物DNA双链断裂修复机制及其在重离子诱变和基因编辑中的作用[J]. 生物技术通报, 2024, 40(7): 55-67. |
| [5] | 陈墨岩, 祝诚. 基于CRISPR/Cas12a的生物传感平台的机制研究及应用[J]. 生物技术通报, 2024, 40(7): 90-98. |
| [6] | 肖怡梦, 杨雯, 程依依, 罗刚. CRISPR-Cas9基因编辑技术及其在家禽中的研究进展[J]. 生物技术通报, 2024, 40(5): 38-47. |
| [7] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [8] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [9] | 朱恬仪, 孔桂美, 焦红梅, 郭停停, 乌日汗, 刘翠翠, 高成凤, 李国才. CRISPR/Cas9介导的adeG基因敲除大肠杆菌细菌模型的建立[J]. 生物技术通报, 2024, 40(2): 55-64. |
| [10] | 高登科, 马白荣, 郭怡莹, 刘薇, 刘田, 靳亚平, 江舟, 陈华涛. 利用CRISPR/Cas9技术构建Quaking敲除的小鼠胚胎成纤维细胞株[J]. 生物技术通报, 2024, 40(2): 65-72. |
| [11] | 张宏民, 龙雯, 劳筱清, 陈雯妍, 商雪梅, 王洪连, 王丽, 粟宏伟, 沈宏萍, 沈宏春. 利用CRISPR/Cas9技术构建Pmepa1基因敲除的TCMK1小鼠肾小管上皮细胞系[J]. 生物技术通报, 2024, 40(2): 73-79. |
| [12] | 周家伟, 武志强. mitoTALENs植物线粒体基因编辑载体的构建方法[J]. 生物技术通报, 2024, 40(10): 172-180. |
| [13] | 皮一飞, 宋新辉, 王淅琳, 李谨谨, 孙长斌, 徐炜. 基于R-loop靶向编辑技术的R-loop功能位点高通量筛选系统[J]. 生物技术通报, 2024, 40(10): 181-190. |
| [14] | 李欣格, 王美霞, 王晨阳, 马桂根, 周常勇, 王亚南, 周焕斌. 基于CRISPR/LanCas9的水稻基因编辑系统的开发和优化[J]. 生物技术通报, 2024, 40(10): 233-242. |
| [15] | 李明坤, 毕美营, 张天航, 吴翔宇, 杨培儒, 应明. UgRNA/Cas9多基因编辑法恢复根际细菌农用功能的研究[J]. 生物技术通报, 2024, 40(10): 275-287. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||