








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 65-72.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0879
高登科1,2( ), 马白荣1,2, 郭怡莹1, 刘薇1,2, 刘田1,2, 靳亚平1,2, 江舟3(
), 马白荣1,2, 郭怡莹1, 刘薇1,2, 刘田1,2, 靳亚平1,2, 江舟3( ), 陈华涛1,2(
), 陈华涛1,2( )
)
收稿日期:2023-09-12
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
陈华涛,男,博士,教授,研究方向:哺乳动物生物钟调控生殖与代谢机制;E-mail: htchen@nwafu.edu.cn;作者简介:高登科,男,博士研究生,研究方向:哺乳动物生物钟调控代谢机制;E-mail: gdk960101@nwafu.edu.cn基金资助:
GAO Deng-ke1,2( ), MA Bai-rong1,2, GUO Yi-ying1, LIU Wei1,2, LIU Tian1,2, JIN Ya-ping1,2, JIANG Zhou3(
), MA Bai-rong1,2, GUO Yi-ying1, LIU Wei1,2, LIU Tian1,2, JIN Ya-ping1,2, JIANG Zhou3( ), CHEN Hua-tao1,2(
), CHEN Hua-tao1,2( )
)
Received:2023-09-12
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】利用CRISPR/Cas9技术构建小鼠胚胎成纤维细胞(NIH3T3)Quaking基因敲除细胞株,并检测Quaking基因对NIH3T3细胞增殖能力的影响。【方法】首先,利用在线网站设计两条靶向作用于Quaking外显子的sgRNA,成功构建了两个分别靶向Quaking基因第1、第2外显子的CRISPR/Cas9重组慢病毒质粒。将构建的Quaking基因CRISPR/Cas9重组慢病毒载体和pcDNA3.1-Quaking过表达质粒共转染至HEK293T细胞中,通过Western blot实验检测Quaking蛋白的敲除效率。其次,将筛选得到的敲除效率高的重组慢病毒质粒(LentiCRISPRv2-sgRNA1)与辅助包装质粒共转染入HEK293T细胞进行慢病毒包装,慢病毒转导NIH3T3细胞后,利用嘌呤霉素筛选阳性单克隆细胞株。最后,通过Western blot及免疫荧光染色鉴定敲除效果。【结果】发现Quaking蛋白在该细胞株中不表达,并测序证实了发生片段敲除。CCK8检测发现,Quaking基因敲除显著抑制了NIH3T3细胞的增殖能力。【结论】本研究首次通过CRISPR/Cas9技术成功构建了小鼠胚胎成纤维细胞(NIH3T3)Quaking基因敲除细胞株,为后续研究Quaking基因在小鼠生理功能调节中的作用机制提供了体外模型基础。
高登科, 马白荣, 郭怡莹, 刘薇, 刘田, 靳亚平, 江舟, 陈华涛. 利用CRISPR/Cas9技术构建Quaking敲除的小鼠胚胎成纤维细胞株[J]. 生物技术通报, 2024, 40(2): 65-72.
GAO Deng-ke, MA Bai-rong, GUO Yi-ying, LIU Wei, LIU Tian, JIN Ya-ping, JIANG Zhou, CHEN Hua-tao. Establishment of Quaking Knockout Mouse Embryonic Fibroblast Cell Line Using CRISPR/Cas9 Technology[J]. Biotechnology Bulletin, 2024, 40(2): 65-72.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| sgRNA1 | CACCGAAGTGCAGAATTGCCTGACG |
| AAACCGTCAGGCAATTCTGCACTTC | |
| sgRNA2 | CACCGGATCTTCAACCACCTCGAG |
| AAACCTCGAGGTGGTTGAAGATCC | |
| Target-F | TTATTTCCCAAAGTGAC |
| Target-R | ATGCCCGAATAGGTT |
表1 序列信息
Table 1 Sequences information
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| sgRNA1 | CACCGAAGTGCAGAATTGCCTGACG |
| AAACCGTCAGGCAATTCTGCACTTC | |
| sgRNA2 | CACCGGATCTTCAACCACCTCGAG |
| AAACCTCGAGGTGGTTGAAGATCC | |
| Target-F | TTATTTCCCAAAGTGAC |
| Target-R | ATGCCCGAATAGGTT |
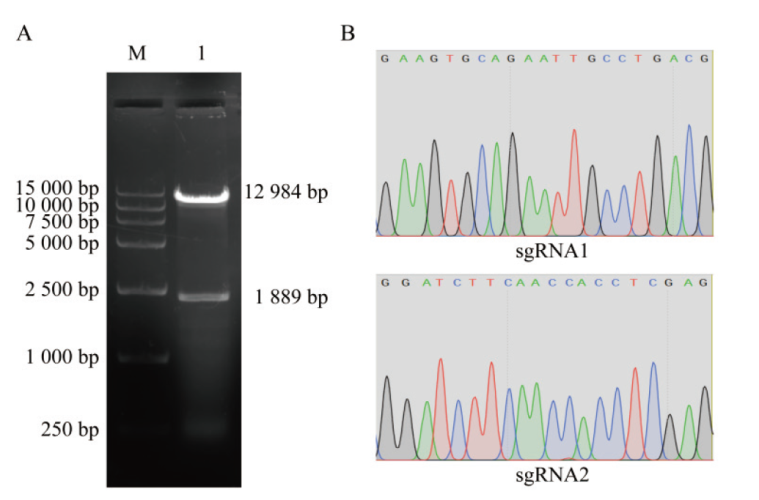
图2 重组质粒的构建与鉴定结果 A:LentiCRISPRv2空载体酶切电泳图,M: marker;1: Esp3I酶切后的LentiCRISPRv2空载体;B:重组质粒LentiCRISPRv2-sgRNA1和LentiCRISPRv2-sgRNA2的测序结果
Fig. 2 Construction and identification of recombinant plasmids A: Electrophoresis of LentiCRISPRv2 empty vector enzyme digestion, M: marker; 1: empty carrier of LentiCRISPRv2 after digestion by Esp3I. B: Sequencing results of recombinant plasmids LentiCRISPRv2-sgRNA1 and LentiCRISPRv2-sgRNA2
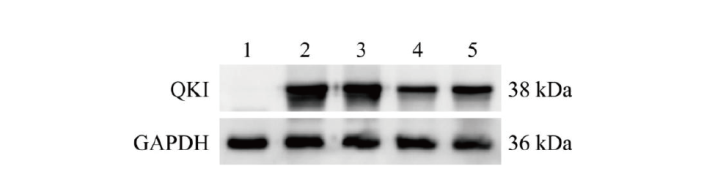
图3 Western blot筛选QKI蛋白敲除效率高的重组质粒 1:未转染的空白细胞对照;2:转染pcDNA3.1-Quaking过表达载体;3:共转染pcDNA3.1-Quaking和LentiCRISPRv2空载体;4:共转染pcDNA3.1-Quaking和LentiCRISPRv2-sgRNA1;5:共转染pcDNA3.1-Quaking和LentiCRISPRv2-sgRNA2
Fig. 3 Screening of recombinant plasmids with high QKI protein knockout efficiency via Western blot 1: Blank control cells without transfection; 2: transfected pcDNA3.1-Quaking overexpression vector; 3: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2 empty vectors; 4: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2-sgRNA1; 5: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2-sgRNA2

图4 Quaking敲除细胞株与野生型细胞株测序结果比对示意图 Wild-type:Quaking野生型细胞株序列;Quaking KO:Quaking敲除细胞株序列;sgRNA用黑色标记,PAM序列用红色标记,蓝色虚线表示删除碱基,括号中的数字表示删除碱基数
Fig. 4 Schematic diagram of comparison between sequencing results of Quaking knockout cell lines and wild-type cell lines Wild-type: Quaking wild-type cell line sequence; Quaking KO: Quaking knockout cell line sequence; sgRNAs are marked in black, PAM sequences in red, dashed blue lines indicate the number of bases removed, and numbers in parentheses indicate the number of bases removed
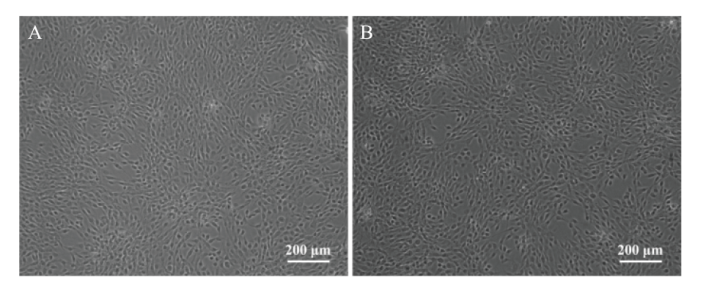
图5 NIH3T3细胞形态图 A:对照组NIH3T3细胞形态 ;B:Quaking基因敲除的NIH3T3形态
Fig. 5 Morphology of NIH3T3 cells A: NIH3T3 cell morphology in control group; B: Quaking gene knockout NIH3T3 morphology
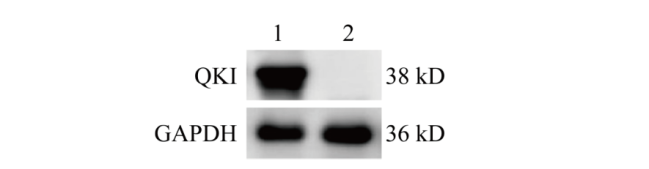
图6 Western blot检测QKI蛋白表达 1:对照组NIH3T3细胞样品;2:Quaking基因敲除的NIH3T3细胞样品
Fig. 6 Detection of QKI protein expression via Western blot 1: NIH3T3 cell samples in control group; 2: NIH3T3 cell samples knocked out by Quaking gene
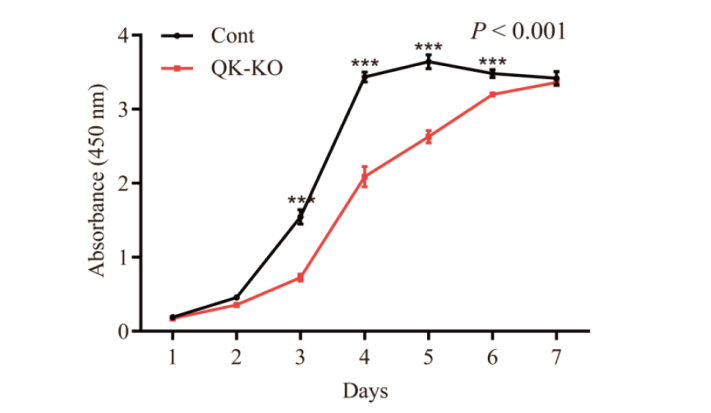
图8 Quaking基因敲除对NIH3T3细胞增殖能力的影响 P < 0.001表示双因素方差分析两组间细胞增殖活力差异极显著;***P < 0.001表示差异极显著
Fig. 8 Effects of Quaking gene knockout on the proliferation ability of NIH3T3 cells P < 0.001 indicates that the cell proliferation activity was significantly different between the two groups in two-factor ANOVA; and ***P < 0.001 indicates a very significant difference
| [1] |
Biedermann B, Hotz HR, Ciosk R. The Quaking family of RNA-binding proteins: coordinators of the cell cycle and differentiation[J]. Cell Cycle, 2010, 9(10): 1929-1933.
pmid: 20495365 |
| [2] |
Ebersole TA, Chen Q, Justice MJ, et al. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins[J]. Nat Genet, 1996, 12(3): 260-265.
doi: 10.1038/ng0396-260 pmid: 8589716 |
| [3] |
Wang YL, Vogel G, Yu ZB, et al. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells[J]. Mol Cell Biol, 2013, 33(6): 1233-1243.
doi: 10.1128/MCB.01604-12 pmid: 23319046 |
| [4] |
Suiko T, Kobayashi K, Aono K, et al. Expression of quaking RNA-binding protein in the adult and developing mouse retina[J]. PLoS One, 2016, 11(5): e0156033.
doi: 10.1371/journal.pone.0156033 URL |
| [5] |
Zhao Y, Zhang G, Wei MY, et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein[J]. Cancer Biol Ther, 2014, 15(1): 108-118.
doi: 10.4161/cbt.26722 pmid: 24153116 |
| [6] | 赵易, 赵庆丽, 马骥, 等. RNA结合蛋白QKI-5对乳腺癌细胞MCF-7增殖的影响研究[J]. 现代生物医学进展, 2017, 17(25): 4816-4819. |
| Zhao Y, Zhao QL, Ma J, et al. Effect of RNA binding protein of QKI-5 on breast cancer cell MCF-7 of proliferation[J]. Prog Mod Biomed, 2017, 17(25): 4816-4819. | |
| [7] | 段小霞, 郭华, 付婷, 等. 震颤同系物-5过表达对卵巢癌细胞A2780增殖和凋亡的影响[J]. 陕西医学杂志, 2022, 51(5): 529-533. |
| Duan XX, Guo H, Fu T, et al. Effect of quaking homolog-5 overexpression on proliferation and apoptosis of ovarian cancer cell line A2780[J]. Shaanxi Med J, 2022, 51(5): 529-533. | |
| [8] |
Wang L, Zhai DS, Ruan BJ, et al. Quaking deficiency amplifies inflammation in experimental endotoxemia via the aryl hydrocarbon receptor/signal transducer and activator of transcription 1-NF-κB pathway[J]. Front Immunol, 2017, 8: 1754.
doi: 10.3389/fimmu.2017.01754 pmid: 29276519 |
| [9] |
Hardy RJ, Loushin CL, Friedrich VL Jr, et al. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice[J]. J Neurosci, 1996, 16(24): 7941-7949.
pmid: 8987822 |
| [10] |
Larocque D, Pilotte J, Chen TP, et al. Nuclear retention of MBP mRNAs in the quaking viable mice[J]. Neuron, 2002, 36(5): 815-829.
doi: 10.1016/s0896-6273(02)01055-3 pmid: 12467586 |
| [11] |
Noveroske JK, Lai LH, Gaussin V, et al. Quaking is essential for blood vessel development[J]. Genesis, 2002, 32(3): 218-230.
pmid: 11892011 |
| [12] |
Bohnsack BL, Lai LH, Northrop JL, et al. Visceral endoderm function is regulated by quaking and required for vascular development[J]. Genesis, 2006, 44(2): 93-104.
pmid: 16470614 |
| [13] | 张杰. NF-κB信号通路对RNA结合蛋白QKI的转录调控及其在骨骼肌中的功能研究[D]. 西安: 第四军医大学, 2009. |
| Zhang J. Transcriptional regulation of RNA binding protein QKI by NF-κB and elucidation of its function in skeletal muscle[D]. Xi'an: The Fourth Military Medical University, 2009. | |
| [14] | 王逢博, 高登科, 李超, 等. 小鼠RNA结合蛋白QKI-5的生物信息学和组织表达分析及其过表达对癌症相关基因表达的影响[J]. 江西农业大学学报, 2023, 45(2): 453-466. |
| Wang FB, Gao DK, Li C, et al. Bioinformatics and tissue expression analysis of mouse RNA binding protein QKI-5 and its overexpression effect on cancer-related genes expression[J]. Acta Agric Univ Jiangxiensis, 2023, 45(2): 453-466. | |
| [15] |
Gao DK, Ma TT, Gao L, et al. Autophagy activation attenuates the circadian clock oscillators in U2OS cells via the ATG5 pathway[J]. Cell Signal, 2023, 101: 110502.
doi: 10.1016/j.cellsig.2022.110502 URL |
| [16] | 刘思远, 易国强, 唐中林, 等. 基于CRISPR/Cas9系统在全基因组范围内筛选功能基因及调控元件研究进展[J]. 遗传, 2020, 42(5): 435-443. |
| Liu SY, Yi GQ, Tang ZL, et al. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements[J]. Hereditas, 2020, 42(5): 435-443. | |
| [17] |
Cong L, Ran FA, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [18] |
Mali P, Yang LH, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826.
doi: 10.1126/science.1232033 pmid: 23287722 |
| [19] |
Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells[J]. eLife, 2013, 2: e00471.
doi: 10.7554/eLife.00471 URL |
| [20] |
Hille F, Richter H, Wong SP, et al. The biology of CRISPR-cas: backward and forward[J]. Cell, 2018, 172(6): 1239-1259.
doi: S0092-8674(17)31383-1 pmid: 29522745 |
| [21] |
Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-cas13[J]. Science, 2017, 358(6366): 1019-1027.
doi: 10.1126/science.aaq0180 pmid: 29070703 |
| [22] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096.
doi: 10.1126/science.1258096 URL |
| [23] |
Kambol R, Gatseva A, Gifford RJ. An endogenous lentivirus in the germline of a rodent[J]. Retrovirology, 2022, 19(1): 30.
doi: 10.1186/s12977-022-00615-2 pmid: 36539757 |
| [24] |
Cockrell AS, Kafri T. Gene delivery by lentivirus vectors[J]. Mol Biotechnol, 2007, 36(3): 184-204.
doi: 10.1007/s12033-007-0010-8 pmid: 17873406 |
| [25] |
You LT, Tong RZ, Li MQ, et al. Advancements and obstacles of CRISPR-Cas9 technology in translational research[J]. Mol Ther Methods Clin Dev, 2019, 13: 359-370.
doi: 10.1016/j.omtm.2019.02.008 URL |
| [26] |
Wang HX, Li MQ, Lee CM, et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery[J]. Chem Rev, 2017, 117(15): 9874-9906.
doi: 10.1021/acs.chemrev.6b00799 URL |
| [27] |
Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases[J]. Nat Rev Genet, 2016, 17(5): 300-312.
doi: 10.1038/nrg.2016.28 pmid: 27087594 |
| [28] |
Wang JZ, Fu X, Fang ZY, et al. QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer[J]. J Mol Cell Biol, 2021, 13(5): 347-360.
doi: 10.1093/jmcb/mjaa063 URL |
| [29] | 梁梦迪. QKI对猪睾丸支持细胞增殖与分泌因子影响的研究[D]. 长春: 吉林大学, 2019. |
| Liang MD. Effects of QKI on the proliferation and secretion factors of pig Sertoli cells[D]. Changchun: Jilin University, 2019. | |
| [30] |
Chen SJ, Niu S, Wang WN, et al. Overexpression of the QKI gene promotes differentiation of goat myoblasts into myotubes[J]. Animals, 2023, 13(4): 725.
doi: 10.3390/ani13040725 URL |
| [1] | 朱恬仪, 孔桂美, 焦红梅, 郭停停, 乌日汗, 刘翠翠, 高成凤, 李国才. CRISPR/Cas9介导的adeG基因敲除大肠杆菌细菌模型的建立[J]. 生物技术通报, 2024, 40(2): 55-64. |
| [2] | 张宏民, 龙雯, 劳筱清, 陈雯妍, 商雪梅, 王洪连, 王丽, 粟宏伟, 沈宏萍, 沈宏春. 利用CRISPR/Cas9技术构建Pmepa1基因敲除的TCMK1小鼠肾小管上皮细胞系[J]. 生物技术通报, 2024, 40(2): 73-79. |
| [3] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [4] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [5] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [6] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [7] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [8] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [9] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [10] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [11] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [12] | 史亚楠, 王德培, 王一川, 周昊, 薛鲜丽. 敲除msn2对米曲霉生长和发酵产曲酸的影响[J]. 生物技术通报, 2022, 38(8): 188-197. |
| [13] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| [14] | Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| [15] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||