








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 85-96.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1066
杨淇( ), 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江(
), 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江( ), 马轩(
), 马轩( )
)
收稿日期:2023-11-13
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
栾维江,男,博士,教授,研究方向:植物分子生物学、水稻功能基因组学;E-mail: skylwj@tjnu.edu.cn;作者简介:杨淇,男,硕士研究生,研究方向:植物分子生物学;E-mail: 18722163533@163.com
基金资助:
YANG Qi( ), WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang(
), WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang( ), MA Xuan(
), MA Xuan( )
)
Received:2023-11-13
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】组蛋白H1对于染色质高级结构的维持和稳定具有重要作用,研究水稻H1对基因表达的影响,为深入理解水稻H1的生物学调控功能提供依据。【方法】通过半定量RT-PCR与RT-qPCR对水稻4个H1基因进行表达分析,利用CRISPR技术创建水稻H1基因编辑植株,鉴定突变体表型,对突变体进行转录组学分析。【结果】水稻4个H1基因的表达比较广谱,在根中的表达较低;在T0代CRISPR突变体筛选过程中发现H1.1-H1.4发生多种突变;在T1代筛选到一株Osh1.1 Osh1.3 Osh1.4纯合三突变体,该三突变体具有多种发育缺陷,成为转录组分析的材料;进一步在T2代得到三突和四突群体,该群体约25%为白化苗,植株生长迟缓,在响应干旱胁迫方面发生缺陷;对三突变体进行转录组学分析,鉴定到1 055个差异表达基因,显著上调的基因约为下调基因的2.5倍,说明H1可能具有抑制基因表达的功能。【结论】在突变体中,光合作用、胁迫响应、氨基酸代谢和RNA代谢等多种途径均发生调控紊乱;其中,核糖体蛋白和光合作用相关基因显著上调,与干旱胁迫相关的脱氢酶基因显著下调。核糖体途径基因过量表达可能造成蛋白质稳态失调,导致植物发育缺陷。
杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96.
YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant[J]. Biotechnology Bulletin, 2024, 40(4): 85-96.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| U-F | CTCCGTTTTACCTGTGGAATCG | CRISPR载体构建 CRISPR vector construction |
| gR-R | CGGAGGAAAATTCCATCCAC | |
| OsH1-T1-gRT1 | TACCTCGGCGTACGGCGGGT | |
| OsH1-T1-U6aT1 | ACCCGCCGTACGCCGAGGTAC | |
| OsH1-T2-gRT1 | ATGATCAAGGAGGCGATCA | |
| OsH1-T2-U6aT1 | TGATCGCCTCCTTGATCATC | |
| PpS-GGL | TTCAGAGGTCTCTCTCGACTAGTATGGAATCGGCAGCAAAGG | |
| PgS-GG2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC | |
| PpS-GG2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC | |
| PgS-GGR | AGCGTGGGTCTCGACCGACGCGTATCCATCCACTCCAAGCTC | |
| OsH1.4F2 | AGCCGGCGAAGGAGAAGAAGAAG | 靶点1检测 Target 1 detection |
| OsH1.4R2 | GGAGAGCTTGAAGGAGTTCTTG | |
| OsH1.3F2 | CTTCGGATAGCGGCAGTAGTCAG | |
| OsH1.3R2 | GCGGCAACTTGTAGGAGTTCTTG | |
| OsH1.2F | AGGCGGAGGGTGAGAAGGAGAAG | 靶点2检测 Target 2 detection |
| OsH1.2R2 | TTCTTCTCCTCGGCAGCCGAC | |
| OsH1.1F2 | TTGGGTTTGGCGGCGTCCTTC | |
| OsH1.1R2 | CAACTTCTCAGCCATGCTCAC | |
| Os04g30420-qRT-F | CAACTTCTCAGCCATGCTCAC | 实时定量PCR RT-qPCR |
| Os04g30420-qRT-R | TCTCAACGACTATCTTGCCGG | |
| Os08g33710-qRT-F | TACACGCTGTCCCAGATCAAG | |
| Os08g33710-qRT-R | TAGAACGCCGGGAACTCGAT | |
| Os03g63950-qRT-F | AGCAGCTAGTGAATGTGGACC | |
| Os03g63950-qRT-R | AGAGTTGGGGATGGTCTCCTT | |
| OsActin-qRT-F | GACTCTGGTGATGGTGTCAGC | |
| OsActin-qRT-R | GGCTGGAAGAGGACCTCAGG | |
| OsH1.1-qRT-F | GTTAAGGCCTCCTACAAGCTCTC | |
| OsH1.1-qRT-R | CTTGGCGACCACCTTCTTCTC | |
| OsH1.2-qRT-F | GAAGGTGAAGGCCTCGTTCAAG | |
| OsH1.2-qRT-R | CTTGTTGGCCTTCTTGGAGATG | |
| OsH1.3-qRT-F | GTGACGAAGACGAAGGCGAC | |
| OsH1.3-qRT-R | CTACTTCTTCGCCTTCCGGG | |
| OsH1.4-qRT-F | CGACCAAGACCAAGATCAAGGT | |
| OsH1.4-qRT-R | CTACTTCATGCTCTTCCTCGCC |
表1 引物信息
Table 1 Primer information
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| U-F | CTCCGTTTTACCTGTGGAATCG | CRISPR载体构建 CRISPR vector construction |
| gR-R | CGGAGGAAAATTCCATCCAC | |
| OsH1-T1-gRT1 | TACCTCGGCGTACGGCGGGT | |
| OsH1-T1-U6aT1 | ACCCGCCGTACGCCGAGGTAC | |
| OsH1-T2-gRT1 | ATGATCAAGGAGGCGATCA | |
| OsH1-T2-U6aT1 | TGATCGCCTCCTTGATCATC | |
| PpS-GGL | TTCAGAGGTCTCTCTCGACTAGTATGGAATCGGCAGCAAAGG | |
| PgS-GG2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC | |
| PpS-GG2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC | |
| PgS-GGR | AGCGTGGGTCTCGACCGACGCGTATCCATCCACTCCAAGCTC | |
| OsH1.4F2 | AGCCGGCGAAGGAGAAGAAGAAG | 靶点1检测 Target 1 detection |
| OsH1.4R2 | GGAGAGCTTGAAGGAGTTCTTG | |
| OsH1.3F2 | CTTCGGATAGCGGCAGTAGTCAG | |
| OsH1.3R2 | GCGGCAACTTGTAGGAGTTCTTG | |
| OsH1.2F | AGGCGGAGGGTGAGAAGGAGAAG | 靶点2检测 Target 2 detection |
| OsH1.2R2 | TTCTTCTCCTCGGCAGCCGAC | |
| OsH1.1F2 | TTGGGTTTGGCGGCGTCCTTC | |
| OsH1.1R2 | CAACTTCTCAGCCATGCTCAC | |
| Os04g30420-qRT-F | CAACTTCTCAGCCATGCTCAC | 实时定量PCR RT-qPCR |
| Os04g30420-qRT-R | TCTCAACGACTATCTTGCCGG | |
| Os08g33710-qRT-F | TACACGCTGTCCCAGATCAAG | |
| Os08g33710-qRT-R | TAGAACGCCGGGAACTCGAT | |
| Os03g63950-qRT-F | AGCAGCTAGTGAATGTGGACC | |
| Os03g63950-qRT-R | AGAGTTGGGGATGGTCTCCTT | |
| OsActin-qRT-F | GACTCTGGTGATGGTGTCAGC | |
| OsActin-qRT-R | GGCTGGAAGAGGACCTCAGG | |
| OsH1.1-qRT-F | GTTAAGGCCTCCTACAAGCTCTC | |
| OsH1.1-qRT-R | CTTGGCGACCACCTTCTTCTC | |
| OsH1.2-qRT-F | GAAGGTGAAGGCCTCGTTCAAG | |
| OsH1.2-qRT-R | CTTGTTGGCCTTCTTGGAGATG | |
| OsH1.3-qRT-F | GTGACGAAGACGAAGGCGAC | |
| OsH1.3-qRT-R | CTACTTCTTCGCCTTCCGGG | |
| OsH1.4-qRT-F | CGACCAAGACCAAGATCAAGGT | |
| OsH1.4-qRT-R | CTACTTCATGCTCTTCCTCGCC |
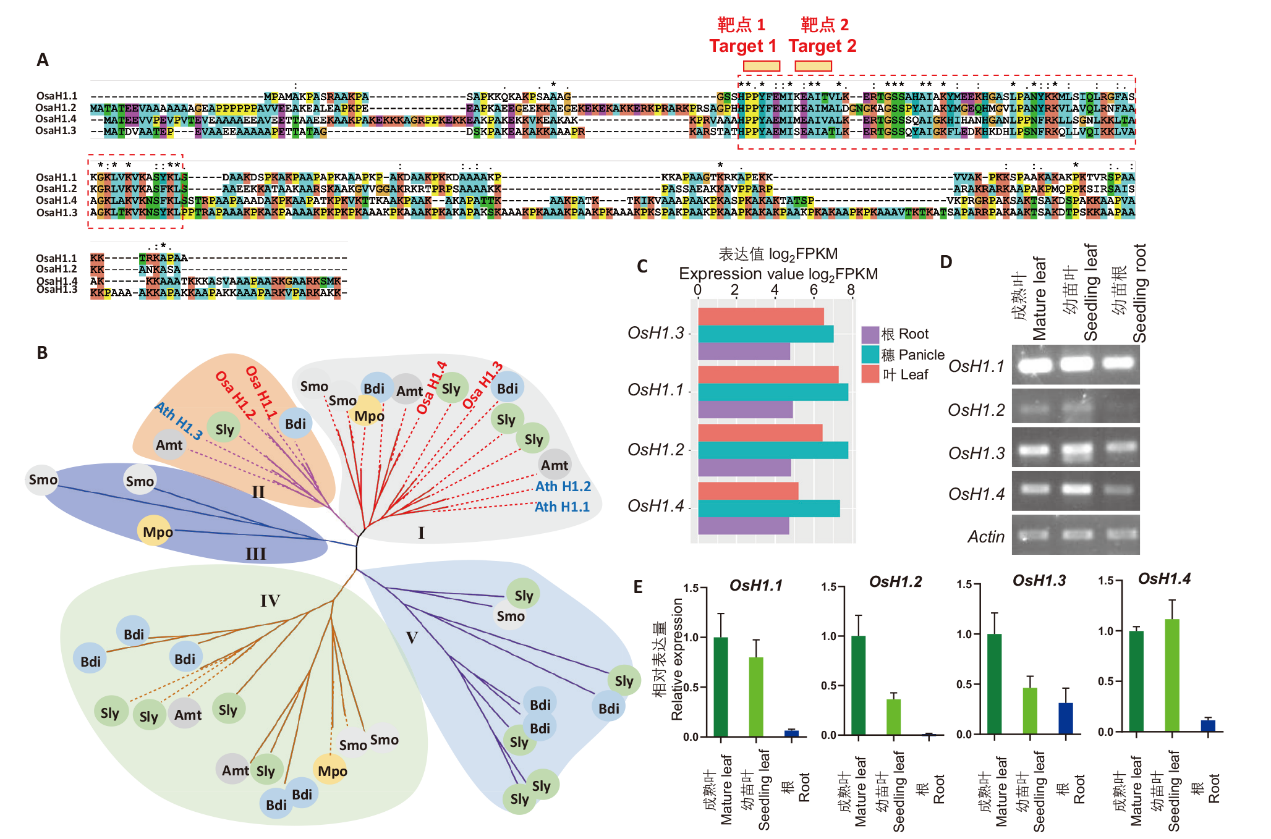
图1 水稻组蛋白H1的序列和表达分析 A:水稻4个组蛋白H1的氨基酸序列比对,红色靶点1和靶点2为CRISPR突变靶点;B:不同植物组蛋白H1的系统进化树(每个分支为1个H1蛋 白,水稻和拟南芥组蛋白H1分别标记红色和蓝色,Mpo:地钱;Smo:卷柏;Atr:无油樟;Ath:拟南芥;Sly:番茄;Bdi:二穗短柄草);C:水稻H1基因的组织特异性表达;D:RT-PCR检测H1在水稻叶和根中的表达;E:RT-qPCR检测H1在水稻叶和根中的表达
Fig. 1 Sequences and expression analysis of rice histone H1 A: Amino acid sequence alignment of four rice histone H1s, red Target 1 and Target 2 are CRISPR mutagenesis sites. B: Phylogenetic tree of histone H1s from different plants(each branch indicates histone H1, and rice and Arabidopsis histone H1s are marked in red and blue, respectively. Species abbreviations: Mpo: Marchantia polymorpha; Smo: Selaginella moellendorffii; Atr: Amborella trichopoda; Ath: Arabidopsis thaliana; Sly: Solanum lycopersicum; Bdi: Brachypodium distachyon). C: Tissue-specific expressions of rice H1 genes. Expression data is derived from RiceSuperPIRdb database. D: RT-PCR detection of H1 expressions in rice shoot and root. E: RT-qPCR detection of H1 expressions in rice shoot and root
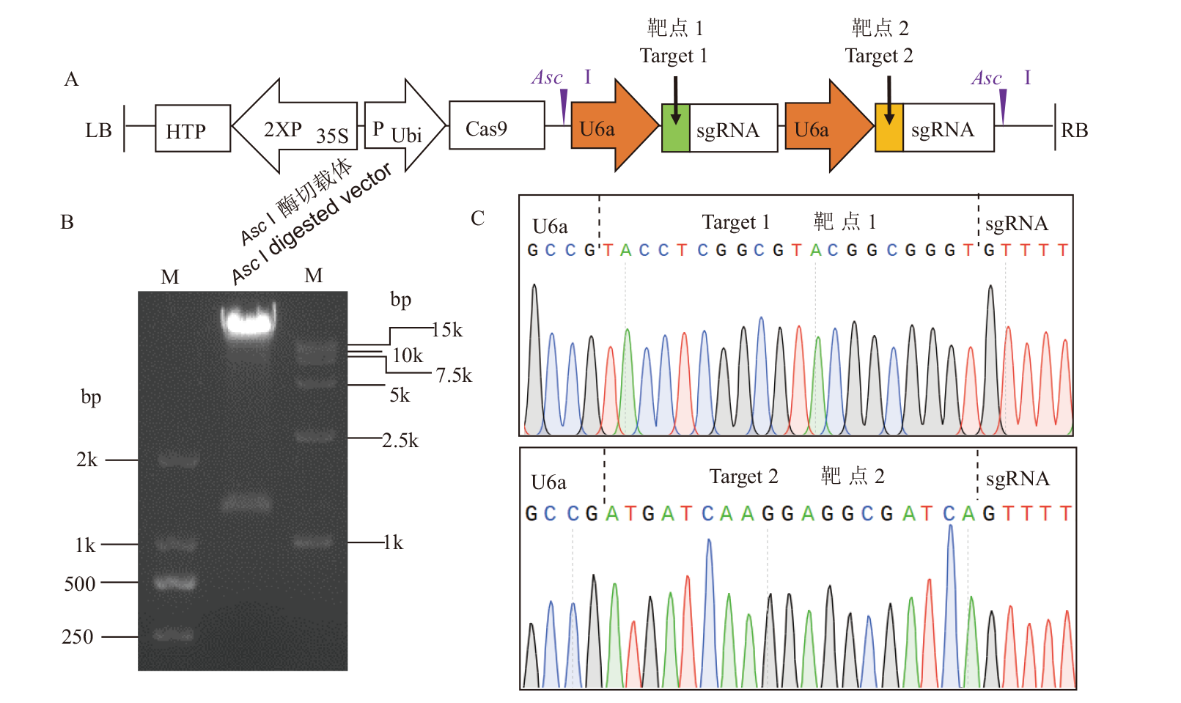
图2 水稻组蛋白H1 CRISPR载体构建 A:含双靶点的pYLCRISPR载体图谱;B:CRISPR载体经Asc I酶切鉴定;M:分子量标记;C:CRISPR载体靶点测序
Fig. 2 Construction of CRISPR vector for rice histone H1 A: Map of pYLCRISPR vector containing two target sites. B: Digestion of CRISPR vector by Asc I. M: Molecular marker. C: Sanger sequencing of CRISPR vector
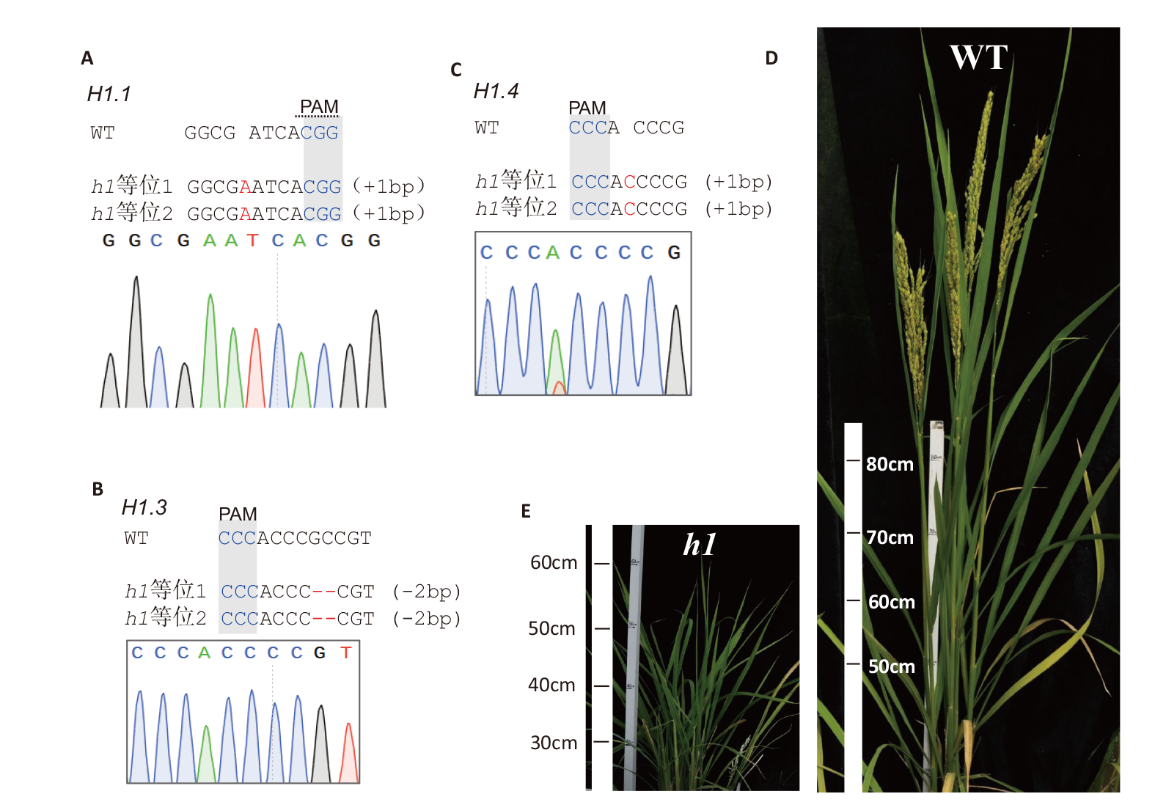
图4 水稻h1三突变体的测序和表型鉴定 A-C:水稻OsH1.1、OsH1.3和OsH1.4的测序鉴定;D-E:水稻野生型和h1三突变体的表型
Fig. 4 Sequencing and phenotypic characterization of rice h1 triple mutant A-C: Sequencing chromatograms of the mutated genes OsH1.1, osH1.3 and osH1.4. D-E: Phenotypes of WT and h1 triple mutant
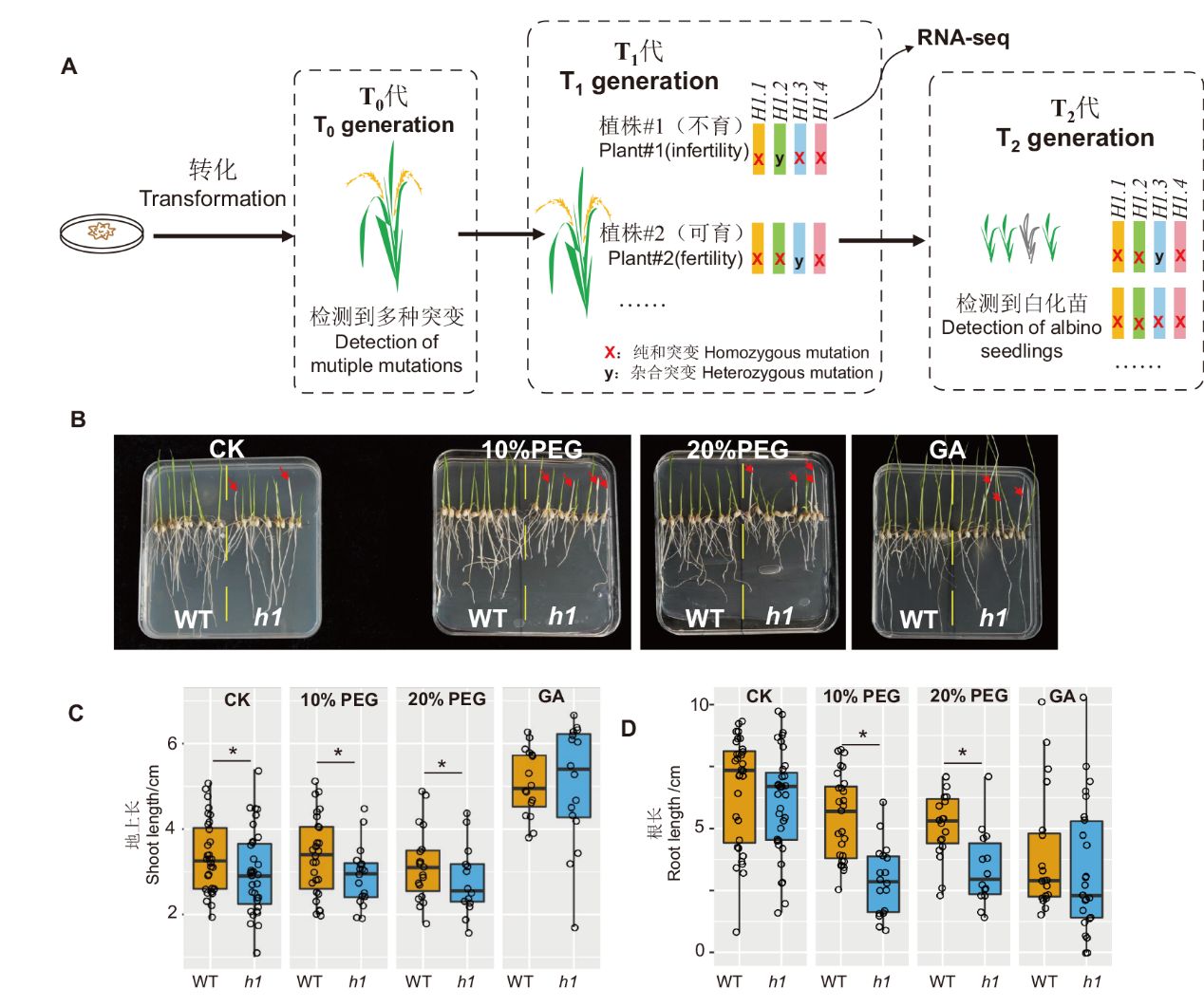
图5 水稻组蛋白h1突变体遗传学及T2代表型分析 A:水稻h1突变体的创建过程;T1代三突植株#1用于RNA-seq,另一株三突植株#2用于传代;B:水稻h1突变体T2代幼苗进行干旱胁迫和施加GA表型鉴定;C:不同处理的叶长统计分析;D:不同处理的根长统计分析。* P<0.05
Fig. 5 Rice h1 mutant genetics and phenotypic analysis of T2 generation A: Construction of rice h1 mutant. The T1 triple plant #1 used for RNA-seq analysis, triple plant #2 used for maintaining mutant alleles. B: Drought stress and GA phenotype identification of rice h1 mutant T2. C: Statistics of shoot lengths of different treatments. D: Statistics of root lengths of different treatments. * P<0.05
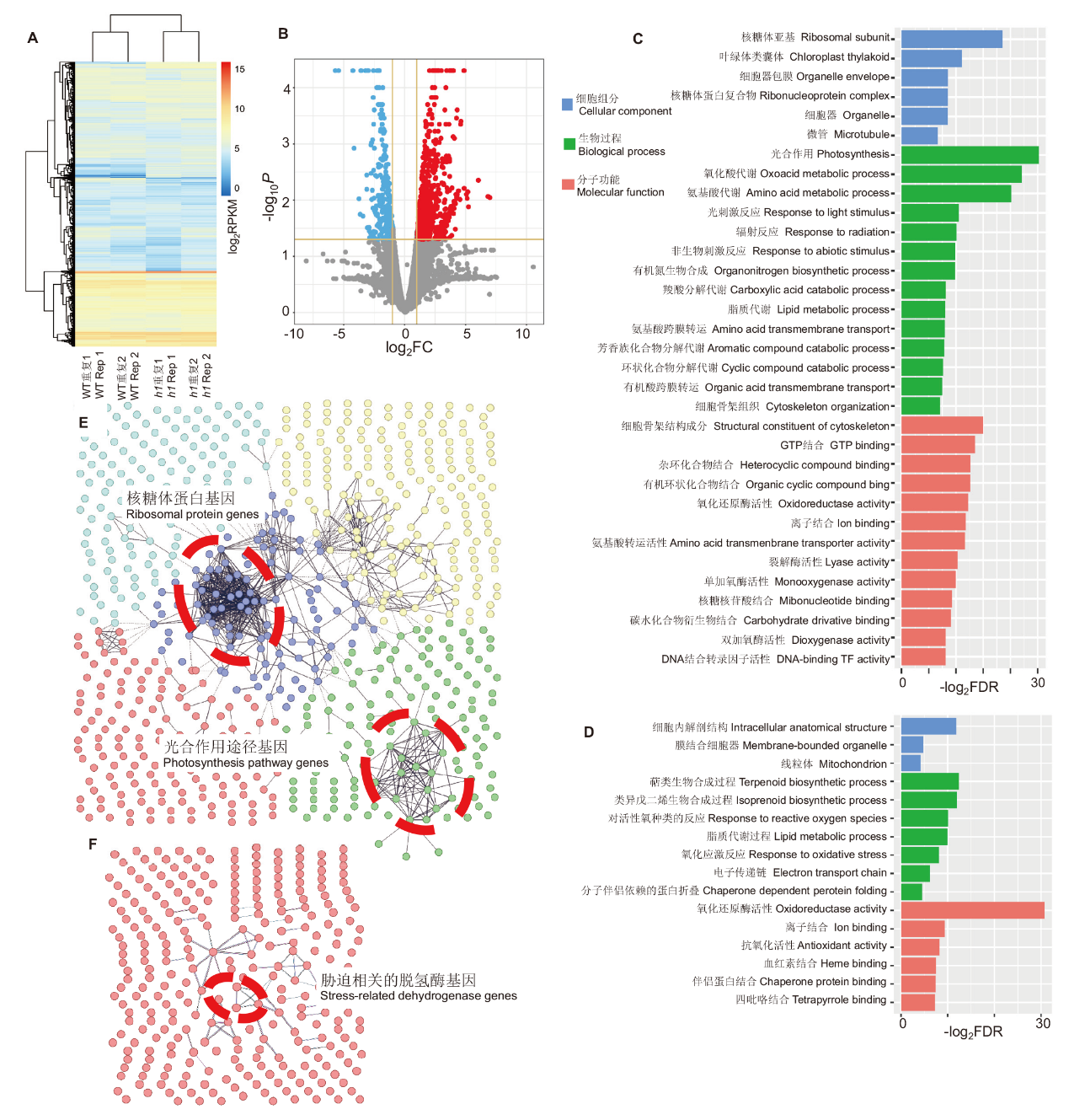
图6 水稻h1三突变体基因表达分析和基因功能分类 A:野生型和h1突变体基因表达比较;B:h1突变体基因表达分析,FC:变化倍数;C:h1突变体上调基因的功能分类,FDR:错误发现率;D:h1突变体下调基因的功能分类;E:上调基因的网络调控分析,红色圈示核糖体蛋白和叶绿体途径基因;F:下调基因的网络调控分析,红色圈示胁迫途径相关的脱氢酶基因
Fig. 6 Gene expression and GO analysis of rice h1 triple mutant A: Comparison of the gene expressions between WT and h1 mutant. B: Gene expression analysis of h1 mutant. FC: Fold-change. C: GO analysis of up-regulated genes in h1 mutant. FDR: False discovery rate. D: GO analysis of down-regulated genes in h1 mutant. E: Network analysis of up-regulated genes. Red circles indicate ribosomal and chloroplast pathway genes. F: Network analysis of down-regulated genes. Red circles indicate stress-related hydrogenase genes
| 基因ID Gene ID | 差异倍数log2值 log2 of fold-change | P值 P value | 功能注释 Functional annotation |
|---|---|---|---|
| LOC_Os03g63950 | 2.701 833 350 | 5.00E-05 | 质体特异性30S核糖体蛋白Plastid-specific 30S ribosomal protein |
| LOC_Os01g14070 | 1.560 583 912 | 0.005 65 | 60S核糖体蛋白L18a-1 60S ribosomal protein L18a-1 |
| LOC_Os03g37970 | 1.212 029 246 | 0.010 8 | 核糖体蛋白L13 Ribosomal protein L13 |
| LOC_Os02g01332 | 1.268 533 286 | 0.011 6 | 核糖体蛋白L6 Ribosomal protein L6 |
| LOC_Os01g01060 | 1.178 811 181 | 0.011 7 | 40S核糖体蛋白S5 40S ribosomal protein S5 |
| LOC_Os07g10720 | 1.276 663 714 | 0.011 7 | 40核糖体蛋白S15a 40S ribosomal protein S15a |
| LOC_Os02g18090 | 1.730 630 752 | 0.013 35 | 线粒体核糖体蛋白L53 Mitochondrial ribosomal protein L53 |
| LOC_Os07g08660 | 1.408 912 387 | 0.014 7 | 40S核糖体蛋白S15 40S ribosomal protein S15 |
| LOC_Os01g10820 | 1.170 916 250 | 0.015 45 | 核糖体蛋白L5 Ribosomal protein L5 |
| LOC_Os04g52361 | 1.177 325 645 | 0.016 1 | 核糖体蛋白S17 Ribosomal protein S17 |
| LOC_Os07g42450 | 1.199 616 805 | 0.016 9 | 核糖体蛋白S2 Ribosomal protein S2 |
| LOC_Os08g13690 | 1.113 868 750 | 0.017 | 60S核糖体蛋白L7 60S ribosomal protein L7 |
| LOC_Os03g60400 | 1.217 299 391 | 0.0175 5 | 40S核糖体蛋白S23 40S ribosomal protein S23 |
| LOC_Os07g10300 | 1.591 065 282 | 0.0188 5 | 线粒体28S核糖体蛋白S29相关 Mitochondrial 28S ribosomal protein S29-related |
| LOC_Os06g19640 | 1.383 070 486 | 0.0202 50 | 线粒体39S核糖体蛋白L46 Mitochondrial 39S ribosomal protein L46 |
| LOC_Os04g28180 | 1.154 726 863 | 0.025 60 | 核糖体蛋白 Ribosomal protein |
| LOC_Os02g30050 | 1.052 697 490 | 0.028 70 | 核糖体蛋白L29 Ribosomal protein L29 |
| LOC_Os07g10660 | 1.148 487 268 | 0.035 30 | 核糖体蛋白Ribosomal protein |
| LOC_Os04g39700 | 1.055 227 902 | 0.040 20 | 60S核糖体蛋白L6 60S ribosomal protein L6 |
| LOC_Os02g32760 | 1.570 591 039 | 0.042 45 | 60S酸性核糖体蛋白 60S acidic ribosomal protein |
| LOC_Os03g48840 | 3.563 924 435 | 0.042 95 | 核糖体蛋白L18p/L5e家族蛋白 Ribosomal L18p/L5e family protein |
| LOC_Os12g38000 | 1.022 593 563 | 0.047 05 | 60S核糖体蛋白L8 60S ribosomal protein L8 |
| LOC_Os08g33820 | 2.001 613 891 | 5.00E-05 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os04g38410 | 2.743 324 248 | 5.00E-05 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os02g36850 | 1.942 328 010 | 0.000 60 | 放氧增强蛋白3 Oxygen-evolving enhancer protein 3 |
| LOC_Os07g37240 | 1.907 040 368 | 0.000 75 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os09g26810 | 1.457 308 201 | 0.001 75 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os07g36080 | 1.656 294 981 | 0.002 45 | 放氧增强蛋白3结构域蛋白 Oxygen-evolving enhancer protein 3 domain protein |
| LOC_Os07g30670 | 1.267 700 635 | 0.014 15 | 2Fe-2S铁硫簇结合域蛋白 2Fe-2S iron-sulfur cluster binding domain protein |
| LOC_Os11g13890 | 1.502 035 534 | 0.014 35 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os12g08770 | 1.118 250 231 | 0.021 50 | 光系统I反应中心亚基 Photosystem I reaction center subunit |
| LOC_Os08g25720 | 1.467 877 984 | 0.023 85 | 焦磷酸-果糖6-磷酸1-磷酸转移酶亚基 Pyrophosphate-fructose 6-phosphate 1-phosphotransferase subunit |
| LOC_Os02g10390 | 1.148 377 518 | 0.029 20 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os06g22060 | 1.526 688 176 | 0.033 45 | 焦磷酸-果糖6-磷酸1-磷酸转移酶亚基 Pyrophosphate-fructose 6-phosphate 1-phosphotransferase subunit |
| LOC_Os06g21590 | 1.221 345 436 | 0.043 55 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
表2 显著上调的核糖体蛋白和光合作用基因
Table 2 Significantly up-regulated ribosomal and photosynthesis genes
| 基因ID Gene ID | 差异倍数log2值 log2 of fold-change | P值 P value | 功能注释 Functional annotation |
|---|---|---|---|
| LOC_Os03g63950 | 2.701 833 350 | 5.00E-05 | 质体特异性30S核糖体蛋白Plastid-specific 30S ribosomal protein |
| LOC_Os01g14070 | 1.560 583 912 | 0.005 65 | 60S核糖体蛋白L18a-1 60S ribosomal protein L18a-1 |
| LOC_Os03g37970 | 1.212 029 246 | 0.010 8 | 核糖体蛋白L13 Ribosomal protein L13 |
| LOC_Os02g01332 | 1.268 533 286 | 0.011 6 | 核糖体蛋白L6 Ribosomal protein L6 |
| LOC_Os01g01060 | 1.178 811 181 | 0.011 7 | 40S核糖体蛋白S5 40S ribosomal protein S5 |
| LOC_Os07g10720 | 1.276 663 714 | 0.011 7 | 40核糖体蛋白S15a 40S ribosomal protein S15a |
| LOC_Os02g18090 | 1.730 630 752 | 0.013 35 | 线粒体核糖体蛋白L53 Mitochondrial ribosomal protein L53 |
| LOC_Os07g08660 | 1.408 912 387 | 0.014 7 | 40S核糖体蛋白S15 40S ribosomal protein S15 |
| LOC_Os01g10820 | 1.170 916 250 | 0.015 45 | 核糖体蛋白L5 Ribosomal protein L5 |
| LOC_Os04g52361 | 1.177 325 645 | 0.016 1 | 核糖体蛋白S17 Ribosomal protein S17 |
| LOC_Os07g42450 | 1.199 616 805 | 0.016 9 | 核糖体蛋白S2 Ribosomal protein S2 |
| LOC_Os08g13690 | 1.113 868 750 | 0.017 | 60S核糖体蛋白L7 60S ribosomal protein L7 |
| LOC_Os03g60400 | 1.217 299 391 | 0.0175 5 | 40S核糖体蛋白S23 40S ribosomal protein S23 |
| LOC_Os07g10300 | 1.591 065 282 | 0.0188 5 | 线粒体28S核糖体蛋白S29相关 Mitochondrial 28S ribosomal protein S29-related |
| LOC_Os06g19640 | 1.383 070 486 | 0.0202 50 | 线粒体39S核糖体蛋白L46 Mitochondrial 39S ribosomal protein L46 |
| LOC_Os04g28180 | 1.154 726 863 | 0.025 60 | 核糖体蛋白 Ribosomal protein |
| LOC_Os02g30050 | 1.052 697 490 | 0.028 70 | 核糖体蛋白L29 Ribosomal protein L29 |
| LOC_Os07g10660 | 1.148 487 268 | 0.035 30 | 核糖体蛋白Ribosomal protein |
| LOC_Os04g39700 | 1.055 227 902 | 0.040 20 | 60S核糖体蛋白L6 60S ribosomal protein L6 |
| LOC_Os02g32760 | 1.570 591 039 | 0.042 45 | 60S酸性核糖体蛋白 60S acidic ribosomal protein |
| LOC_Os03g48840 | 3.563 924 435 | 0.042 95 | 核糖体蛋白L18p/L5e家族蛋白 Ribosomal L18p/L5e family protein |
| LOC_Os12g38000 | 1.022 593 563 | 0.047 05 | 60S核糖体蛋白L8 60S ribosomal protein L8 |
| LOC_Os08g33820 | 2.001 613 891 | 5.00E-05 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os04g38410 | 2.743 324 248 | 5.00E-05 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os02g36850 | 1.942 328 010 | 0.000 60 | 放氧增强蛋白3 Oxygen-evolving enhancer protein 3 |
| LOC_Os07g37240 | 1.907 040 368 | 0.000 75 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os09g26810 | 1.457 308 201 | 0.001 75 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os07g36080 | 1.656 294 981 | 0.002 45 | 放氧增强蛋白3结构域蛋白 Oxygen-evolving enhancer protein 3 domain protein |
| LOC_Os07g30670 | 1.267 700 635 | 0.014 15 | 2Fe-2S铁硫簇结合域蛋白 2Fe-2S iron-sulfur cluster binding domain protein |
| LOC_Os11g13890 | 1.502 035 534 | 0.014 35 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os12g08770 | 1.118 250 231 | 0.021 50 | 光系统I反应中心亚基 Photosystem I reaction center subunit |
| LOC_Os08g25720 | 1.467 877 984 | 0.023 85 | 焦磷酸-果糖6-磷酸1-磷酸转移酶亚基 Pyrophosphate-fructose 6-phosphate 1-phosphotransferase subunit |
| LOC_Os02g10390 | 1.148 377 518 | 0.029 20 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
| LOC_Os06g22060 | 1.526 688 176 | 0.033 45 | 焦磷酸-果糖6-磷酸1-磷酸转移酶亚基 Pyrophosphate-fructose 6-phosphate 1-phosphotransferase subunit |
| LOC_Os06g21590 | 1.221 345 436 | 0.043 55 | 叶绿素A-B结合蛋白 Chlorophyll A-B binding protein |
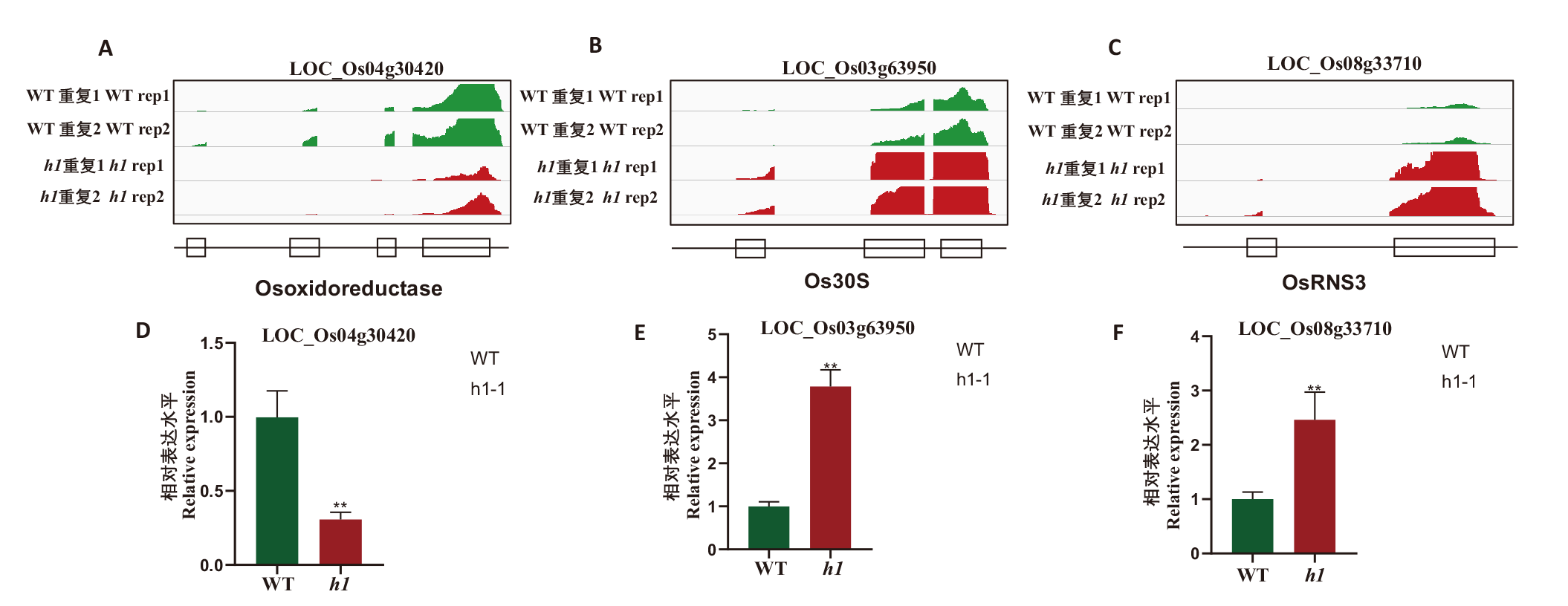
图7 水稻3个基因在h1突变体中的表达发生紊乱 A-C:转录组测序峰图,每个样品两次重复;D-F:RT-qPCR验证。** P<0.01
Fig. 7 Expressions of three genes are disrupted in rice h1 triple mutant A-C: Transcriptomic sequencing peaks, there are 2 replicates for each sample. D-F: RT-qPCR validation. ** P < 0.01
| [1] |
Chen RZ, Deng YW, Ding YL, et al. Rice functional genomics: decades’ efforts and roads ahead[J]. Sci China Life Sci, 2022, 65(1): 33-92.
doi: 10.1007/s11427-021-2024-0 |
| [2] |
Xie L, Liu MH, Zhao L, et al. RiceENCODE: A comprehensive epigenomic database as a rice Encyclopedia of DNA Elements[J]. Mol Plant, 2021, 14(10): 1604-1606.
doi: 10.1016/j.molp.2021.08.018 URL |
| [3] |
Tang SJ, Yang C, Wang D, et al. Targeted DNA demethylation produces heritable epialleles in rice[J]. Sci China Life Sci, 2022, 65(4): 753-756.
doi: 10.1007/s11427-021-1974-7 |
| [4] |
Kornberg RD. Chromatin structure: A repeating unit of histones and DNA[J]. Science, 1974, 184(4139): 868-871.
doi: 10.1126/science.184.4139.868 pmid: 4825889 |
| [5] |
Khorasanizadeh S. The nucleosome: From genomic organization to genomic regulation[J]. Cell, 2004, 116(2): 259-272.
pmid: 14744436 |
| [6] |
Izzo A, Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics[J]. Biochim Biophys Acta, 2016, 1859(3): 486-495.
doi: 10.1016/j.bbagrm.2015.09.003 pmid: 26348411 |
| [7] | Bednar J, Garcia-saez I, Boopathi R, et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1[J]. Mol Cell, 2017, 66(5): 729. |
| [8] |
Fyodorov DV, Zhou BR, Skoultchi AI, et al. Emerging roles of linker histones in regulating chromatin structure and function[J]. Nat Rev Mol Cell Biol, 2018, 19(3): 192-206.
doi: 10.1038/nrm.2017.94 URL |
| [9] |
Jerzmanowski A, Przewłoka M, Grasser KD. Linker histones and HMG1 proteins of higher plants[J]. Plant Biol, 2000, 2(6): 586-597.
doi: 10.1055/s-2000-16648 URL |
| [10] |
Talbert PB, Ahmad K, Almouzni G, et al. A unified phylogeny-based nomenclature for histone variants[J]. Epigenetics Chromatin, 2012, 5: 7.
doi: 10.1186/1756-8935-5-7 pmid: 22650316 |
| [11] |
Rutowicz K, Puzio M, Halibart-puzio J, et al. A Specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis[J]. Plant Physiol, 2015, 169(3): 2080-2101.
doi: 10.1104/pp.15.00493 pmid: 26351307 |
| [12] |
Cohen A, Plant AL, Moses MS, et al. Organ-specific and environmentally regulated expression of two abscisic acid-induced genes of tomato 1: Nucleotide sequence and analysis of the corresponding cDNAs[J]. Plant Physiol, 1991, 97(4): 1367-1374.
doi: 10.1104/pp.97.4.1367 pmid: 16668558 |
| [13] |
Wei T, O'connell MA. Structure and characterization of a putative drought-inducible H1 histone gene[J]. Plant mol biol, 1996, 30 (2): 255-268.
doi: 10.1007/BF00020112 pmid: 8616250 |
| [14] |
Przewloka MR, Wierzbicki AT, Ślusarczyk J, et al. The “drought-inducible” histone H1s of tobacco play no role in male sterility linked to alterations in H1 variants[J]. Planta, 2002, 215(3): 371-379.
doi: 10.1007/s00425-002-0758-9 pmid: 12111217 |
| [15] |
Ascenzi R, Gantt JS. A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants[J]. Plant Mol Biol, 1997, 34(4): 629-641.
pmid: 9247544 |
| [16] |
Sheikh AH, Nawaz K, Tabassum N, et al. Linker histone H1 modulates defense priming and immunity in plants[J]. Nucleic Acids Res, 2023, 51(9): 4252-65.
doi: 10.1093/nar/gkad106 URL |
| [17] |
Scippa GS, Di Michele M, Onelli E, et al. The histone-like protein H1-S and the response of tomato leaves to water deficit[J]. J Exp Bot, 2004, 55(394): 99-109.
doi: 10.1093/jxb/erh022 pmid: 14645393 |
| [18] |
Rea M, Zheng W, Chen M, et al. Histone H1 affects gene imprinting and DNA methylation in Arabidopsis[J]. Plant J, 2012, 71(5): 776-786.
doi: 10.1111/tpj.2012.71.issue-5 URL |
| [19] | Rutowicz K, Lirski M, Mermaz B, et al. Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis[J]. Genome Biol, 2019, 20(1): 157. |
| [20] |
Choi J, Lyons DB, Zilberman D. Histone H1 prevents non-CG methylation-mediated small RNA biogenesis in Arabidopsis heterochromatin[J]. elife, 2021, 10: e72676.
doi: 10.7554/eLife.72676 URL |
| [21] |
Choi J, Lyons DB, Kim MY, et al. DNA Methylation and histone H1 jointly repress transposable elements and aberrant intragenic transcripts[J]. Mol Cell, 2020, 77(2): 310-323. e7.
doi: S1097-2765(19)30789-0 pmid: 31732458 |
| [22] | Bourguet P, Picard CL, Yelagandula R, et al. The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation[J]. Nat Commun, 2021, 12(1): 2683. |
| [23] |
He SB, Vickers M, Zhang JY, et al. Natural depletion of histone H1 in sex cells causes DNA demethylation, heterochromatin decondensation and transposon activation[J]. eLife, 2019, 8: e42530.
doi: 10.7554/eLife.42530 URL |
| [24] |
Hu Y, Lai Y. Identification and expression analysis of rice histone genes[J]. Plant Physiol Biochem, 2015, 86: 55-65.
doi: 10.1016/j.plaphy.2014.11.012 URL |
| [25] | Wan JL, Zhang J, Zan XF, et al. Overexpression of rice histone H1 gene reduces tolerance to cold and heat stress[J]. Plants(Basel), 2023, 12(13): 2408. |
| [26] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8): 1274-1284.
doi: 10.1016/j.molp.2015.04.007 pmid: 25917172 |
| [27] | Tadini L, Jeran N, Domingo G, et al. Perturbation of protein homeostasis brings plastids at the crossroad between repair and dismantling[J]. PLoS Genet, 2023, 19(7): e1010344. |
| [28] |
Hedden P. Gibberellin metabolism and its regulation[J]. J Plant Growth Regul, 2001, 20(4): 317-318.
doi: 10.1007/s003440010039 pmid: 11986757 |
| [29] |
Wilkins O, Hafemeister C, Plessis A, et al. EGRINs(environmental gene regulatory influence networks)in rice that function in the response to water deficit, high temperature, and agricultural environments[J]. Plant Cell, 2016, 28(10): 2365-2384.
doi: 10.1105/tpc.16.00158 URL |
| [30] |
Gho YS, Choi H, Moon S, et al. Phosphate-starvation-inducible S-like RNase genes in rice are involved in phosphate source recycling by RNA decay[J]. Front Plant Sci, 2020, 11: 585-561.
doi: 10.3389/fpls.2020.00585 URL |
| [31] |
Liu X, Song LL, Zhang H, et al. Rice ubiquitin-conjugating enzyme OsUBC26 is essential for immunity to the blast fungus Magnaporthe oryzae[J]. Mol Plant Pathol, 2021, 22(12): 1613-1623.
doi: 10.1111/mpp.13132 URL |
| [32] |
MacIntosh GC, Hillwig MS, Meyer A, et al. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants[J]. Mol Genet Genomics, 2010, 283(4): 381-396.
doi: 10.1007/s00438-010-0524-9 pmid: 20182746 |
| [1] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [2] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [3] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [4] | 朱恬仪, 孔桂美, 焦红梅, 郭停停, 乌日汗, 刘翠翠, 高成凤, 李国才. CRISPR/Cas9介导的adeG基因敲除大肠杆菌细菌模型的建立[J]. 生物技术通报, 2024, 40(2): 55-64. |
| [5] | 高登科, 马白荣, 郭怡莹, 刘薇, 刘田, 靳亚平, 江舟, 陈华涛. 利用CRISPR/Cas9技术构建Quaking敲除的小鼠胚胎成纤维细胞株[J]. 生物技术通报, 2024, 40(2): 65-72. |
| [6] | 张宏民, 龙雯, 劳筱清, 陈雯妍, 商雪梅, 王洪连, 王丽, 粟宏伟, 沈宏萍, 沈宏春. 利用CRISPR/Cas9技术构建Pmepa1基因敲除的TCMK1小鼠肾小管上皮细胞系[J]. 生物技术通报, 2024, 40(2): 73-79. |
| [7] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [8] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| [9] | 林鑫焱, 张传忠, 戴兵, 王馨珩, 刘剑锋, 温丽, 徐兴健, 方军. 水稻穗发芽遗传与分子机制的研究进展[J]. 生物技术通报, 2024, 40(1): 24-31. |
| [10] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [11] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [12] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [13] | 刘佳慧, 刘叶, 花尔并, 王猛. 谷氨酸棒杆菌中胞嘧啶碱基编辑工具的PAM拓展[J]. 生物技术通报, 2023, 39(9): 49-57. |
| [14] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [15] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||